
- DEPOD Home
- About DEPOD
- User Manual
- Human Phosphatases
- Protein Substrates
- Non-Protein Substrates
- Pathway Mapping
- Dephosphorylation Network
- ID Mapping
- Download
- Acknowledgement
- Contact
- Imprint
- Links
- Ensembl
- Entrez Gene
- HGNC
- QuickGO
- Gene Expression Atlas
- OMIM
- UniProt
- IntEnz
- BRENDA
- LocDB
- M-CSA
- InterPro
- Pfam
- CATH
- Gene3D
- PDB
- PDBe
- DrugPort
- ModBase
- SwissModel
- ChEMBL
- Phospho.ELM
- PhosphoSitePlus
- NetworKIN
- HPRD
- PSICQUIC
- STRING
- IntAct
- MINT
- KEGG Pathway
- Reactome Pathway
- Pathway Commons
- PubMed
- ArchSchema
- Cytoscape Web
- Europe PMC
- EKPD
- SwissLipids
 Statistics
Statistics
194 phosphatases have substrate data;
--------------------------------
336 protein substrates;
83 non-protein substrates;
1215 dephosphorylation interactions;
--------------------------------
299 KEGG pathways;
876 Reactome pathways;
--------------------------------
last scientific update:
11 Mar, 2019

Top
PGAM1
| Gene Name | PGAM1 (QuickGO) | (A quick tutorial to explore the interctive visulaization) |
| Synonyms | PGAM1, PGAMA, CDABP0006 | |
| Protein Name | PGAM1 |
|
| Alternative Name(s) |
Phosphoglycerate mutase 1;5.4.2.11;5.4.2.4;BPG-dependent PGAM 1;Phosphoglycerate mutase isozyme B;PGAM-B; | |
| EntrezGene ID | 5223 (Comparitive Toxicogenomics) | |
| UniProt AC (Human) | P18669 (protein sequence) | |
| Enzyme Class | EC 5.4.2.11 5.4.2.4 (BRENDA | |
| Molecular Weight | 28804 Dalton | |
| Protein Length | 254 amino acids (AA) | |
| Genome Browsers | NCBI | ENSG00000171314 (Ensembl) | UCSC | |
| Crosslinking annotations | Query our ID-mapping table | |
| Orthologues | Quest For Orthologues (QFO) | GeneTree | |
| Classification | Superfamily: histidine phosphatase (HP) --> Family: PGAM | Historic class: Phosphoglycerate mutase | CATH ID: 3.40.50.1240 | SCOP Fold: HP | |
| Phosphatase activity | active | Catalytic signature motif: unknown | |
| Phosphorylation Network | Visualize | |
| Domain organization, Expression, Diseases | (show / hide) |
| Protein Domain | InterPro | Pfam | SMART | 3D Structure | PDB | DrugPort | ModBase | SwissModel |
| Polypeptide organization (Pfam) | No Pfam domain found | ||
| Catalytic Site | Mechanism and Catalytic Site Atlas | ||
| Gene Expression | Gene Expression Atlas | Diseases | OMIM | DisGeNet | ClinVar |
| Protein-protein interactions | STRING | IntAct | MINT | BioGRID | ||
| Phosphosites | Phospho.ELM | PhosphoSitePlus | ||
| Localization, Function, Catalytic activity and Sequence | (show / hide) |
| Localization (UniProt annotation) | |||
| N/A | |||
| Function (UniProt annotation) | |||
| Interconversion of 3- and 2-phosphoglycerate with 2,3-bisphosphoglycerate as the primer of the reaction Can alsocatalyze the reaction of EC 5424 (synthase), but with a reducedactivity | |||
| Catalytic Activity (UniProt annotation) | |||
| 2-phospho-D-glycerate = 3-phospho-D-glycerate 3-phospho-D-glyceroyl phosphate = 2,3-bisphospho-D-glycerate | |||
| Protein Sequence | |||
| MAAYKLVLIRHGESAWNLENRFSGWYDADLSPAGHEEAKRGGQALRDAGYEFDICFTSVQKRAIRTLWTVLDAIDQMWLP VVRTWRLNERHYGGLTGLNKAETAAKHGEAQVKIWRRSYDVPPPPMEPDHPFYSNISKDRRYADLTEDQLPSCESLKDTI ARALPFWNEEIVPQIKEGKRVLIAAHGNSLRGIVKHLEGLSEEAIMELNLPTGIPIVYELDKNLKPIKPMQFLGDEETVR KAMEAVAAQGKAKK |
| Motif information from Eukaryotic Linear Motif atlas (ELM) | (show / hide) |
| Gene Ontology (P: Process; F: Function and C: Component terms) | (show / hide) | |
| GO:0004082 | F:bisphosphoglycerate mutase activity |
| GO:0004619 | F:phosphoglycerate mutase activity |
| GO:0005576 | C:extracellular region |
| GO:0005634 | C:nucleus |
| GO:0005737 | C:cytoplasm |
| GO:0005829 | C:cytosol |
| GO:0006094 | P:gluconeogenesis |
| GO:0006096 | P:glycolytic process |
| GO:0006110 | P:regulation of glycolytic process |
| GO:0016020 | C:membrane |
| GO:0016787 | F:hydrolase activity |
| GO:0019901 | F:protein kinase binding |
| GO:0034774 | C:secretory granule lumen |
| GO:0043209 | C:myelin sheath |
| GO:0043312 | P:neutrophil degranulation |
| GO:0043456 | P:regulation of pentose-phosphate shunt |
| GO:0045730 | P:respiratory burst |
| GO:0046538 | F:2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity |
| GO:0061621 | P:canonical glycolysis |
| GO:0070062 | C:extracellular exosome |
| GO:1904813 | C:ficolin-1-rich granule lumen |
[Feedback|Contribution]
| Substrate ChEBI-ID | Substrate KEGG ID | DephosphoSite | Bioassay Type | PubMed ID | View in Europe PMC | Reliability of the interaction | Molecule |
| CHEBI:17720 | C01159 | N/A | N/A | UniProt, | Europe PMC |  | 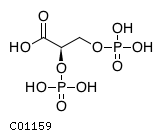 |
| Pathway ID | Pathway Name | Pathway Description (KEGG) |
| hsa00010 | Glycolysis / Gluconeogenesis | Glycolysis is the process of converting glucose into pyruvate and generating small amounts of ATP (energy) and NADH (reducing power). It is a central pathway that produces important precursor metabolites: six-carbon compounds of glucose-6P and fructose-6P and three-carbon compounds of glycerone-P, glyceraldehyde-3P, glycerate-3P, phosphoenolpyruvate, and pyruvate [MD:M00001]. Acetyl-CoA, another important precursor metabolite, is produced by oxidative decarboxylation of pyruvate [MD:M00307]. When the enzyme genes of this pathway are examined in completely sequenced genomes, the reaction steps of three-carbon compounds from glycerone-P to pyruvate form a conserved core module [MD:M00002], which is found in almost all organisms and which sometimes contains operon structures in bacterial genomes. Gluconeogenesis is a synthesis pathway of glucose from noncarbohydrate precursors. It is essentially a reversal of glycolysis with minor variations of alternative paths [MD:M00003]. |
| hsa00260 | Glycine, serine and threonine metabolism | Serine is derived from 3-phospho-D-glycerate, an intermediate of glycolysis [MD:M00020], and glycine is derived from serine. Threonine is an essential amino acid, which animals cannot synthesize. In bacteria and plants, threonine is derived from aspartate [MD:M00018]. |
| hsa01100 | Metabolic pathways | |
| hsa01200 | Carbon metabolism | Carbon metabolism is the most basic aspect of life. This map presents an overall view of central carbon metabolism, where the number of carbons is shown for each compound denoted by a circle, excluding a cofactor (CoA, CoM, THF, or THMPT) that is replaced by an asterisk. The map contains carbon utilization pathways of glycolysis (map00010), pentose phosphate pathway (map00030), and citrate cycle (map00020), and six known carbon fixation pathways (map00710 and map00720) as well as some pathways of methane metabolism (map00680). The six carbon fixation pathways are: (1) reductive pentose phosphate cycle (Calvin cycle) in plants and cyanobacteria that perform oxygenic photosynthesis, (2) reductive citrate cycle in photosynthetic green sulfur bacteria and some chemolithoautotrophs, (3) 3-hydroxypropionate bi-cycle in photosynthetic green nonsulfur bacteria, two variants of 4-hydroxybutyrate pathways in Crenarchaeota called (4) hydroxypropionate-hydroxybutyrate cycle and (5) dicarboxylate-hydroxybutyrate cycle, and (6) reductive acetyl-CoA pathway in methanogenic bacteria. |
| hsa01230 | Biosynthesis of amino acids | This map presents a modular architecture of the biosynthesis pathways of twenty amino acids, which may be viewed as consisting of the core part and its extensions. The core part is the KEGG module for conversion of three-carbon compounds from glyceraldehyde-3P to pyruvate [MD:M00002], together with the pathways around serine and glycine. This KEGG module is the most conserved one in the KEGG MODULE database and is found in almost all the completely sequenced genomes. The extensions are the pathways containing the reaction modules RM001, RM033, RM032, and RM002 for biosynthesis of branched-chain amino acids (left) and basic amino acids (bottom), and the pathways for biosynthesis of histidine and aromatic amino acids (top right). It is interesting to note that the so-called essential amino acids that cannot be synthesized in human and other organisms generally appear in these extensions. Furthermore, the bottom extension of basic amino acids appears to be most divergent containing multiple pathways for lysine biosynthesis and multiple gene sets for arginine biosynthesis. |
| hsa04922 | Glucagon signaling pathway | Glucagon is conventionally regarded as a counterregulatory hormone for insulin and plays a critical anti-hypoglycemic role by maintaining glucose homeostasis in both animals and humans. To increase blood glucose, glucagon promotes hepatic glucose output by increasing glycogenolysis and gluconeogenesis and by decreasing glycogenesis and glycolysis in a concerted fashion via multiple mechanisms. Glucagon also stimulates hepatic mitochondrial beta-oxidation to supply energy for glucose production. Glucagon performs its main effect via activation of adenylate cyclase. The adenylate-cyclase-derived cAMP activates protein kinase A (PKA), which then phosphorylates downstream targets, such as cAMP response element binding protein (CREB) and the bifunctional enzyme 6-phosphofructo-2-kinase/ fructose-2,6-bisphosphatase (one of the isoforms being PFK/FBPase 1, encoded by PFKFB1). |
| hsa05230 | Central carbon metabolism in cancer | Malignant transformation of cells requires specific adaptations of cellular metabolism to support growth and survival. In the early twentieth century, Otto Warburg established that there are fundamental differences in the central metabolic pathways operating in malignant tissue. He showed that cancer cells consume a large amount of glucose, maintain high rate of glycolysis and convert a majority of glucose into lactic acid even under normal oxygen concentrations (Warburg's Effects). More recently, it has been recognized that the 'Warburg effect' encompasses a similarly increased utilization of glutamine. From the intermediate molecules provided by enhanced glycolysis and glutaminolysis, cancer cells synthesize most of the macromolecules required for the duplication of their biomass and genome. These cancer-specific alterations represent a major consequence of genetic mutations and the ensuing changes of signalling pathways in cancer cells. Three transcription factors, c-MYC, HIF-1 and p53, are key regulators and coordinate regulation of cancer metabolism in different ways, and many other oncogenes and tumor suppressor genes cluster along the signaling pathways that regulate c-MYC, HIF-1 and p53. |
| Pathway ID | Pathway Name | Pathway Description (KEGG) |
| R-HSA-6798695 | Neutrophil degranulation. | Neutrophils are the most abundant leukocytes (white blood cells), indispensable in defending the body against invading microorganisms. In response to infection, neutrophils leave the circulation and migrate towards the inflammatory focus. They contain several subsets of granules that are mobilized to fuse with the cell membrane or phagosomal membrane, resulting in the exocytosis or exposure of membrane proteins. Traditionally, neutrophil granule constituents are described as antimicrobial or proteolytic, but granules also introduce membrane proteins to the cell surface, changing how the neutrophil responds to its environment (Borregaard et al. 2007). Primed neutrophils actively secrete cytokines and other inflammatory mediators and can present antigens via MHC II, stimulating T-cells (Wright et al. 2010).Granules form during neutrophil differentiation. Granule subtypes can be distinguished by their content but overlap in structure and composition. The differences are believed to be a consequence of changing protein expression and differential timing of granule formation during the terminal processes of neutrophil differentiation, rather than sorting (Le Cabec et al. 1996). The classical granule subsets are Azurophil or primary granules (AG), secondary granules (SG) and gelatinase granules (GG). Neutrophils also contain exocytosable storage cell organelles, storage vesicles (SV), formed by endocytosis they contain many cell-surface markers and extracellular, plasma proteins (Borregaard et al. 1992). Ficolin-1-rich granules (FG) are like GGs highly exocytosable but gelatinase-poor (Rorvig et al. 2009) |
| R-HSA-70171 | Glycolysis. | The reactions of glycolysis (e.g., van Wijk and van Solinge 2005) convert glucose 6-phosphate to pyruvate. The entire process is cytosolic. Glucose 6-phosphate is reversibly isomerized to form fructose 6-phosphate. Phosphofructokinase 1 catalyzes the physiologically irreversible phosphorylation of fructose 6-phosphate to form fructose 1,6-bisphosphate. In six reversible reactions, fructose 1,6-bisphosphate is converted to two molecules of phosphoenolpyruvate and two molecules of NAD+ are reduced to NADH + H+. Each molecule of phosphoenolpyruvate reacts with ADP to form ATP and pyruvate in a physiologically irreversible reaction. Under aerobic conditions the NADH +H+ can be reoxidized to NAD+ via electron transport to yield additional ATP, while under anaerobic conditions or in cells lacking mitochondria NAD+ can be regenerated via the reduction of pyruvate to lactate |
| R-HSA-70263 | Gluconeogenesis. | The reactions of gluconeogenesis convert mitochondrial pyruvate to cytosolic glucose 6-phosphate which in turn can be hydrolyzed to glucose and exported from the cell. Gluconeogenesis is confined to cells of the liver and kidney and enables glucose synthesis from molecules such as lactate and alanine and other amino acids when exogenous glucose is not available (reviewed, e.g., by Gerich 1993). The process of gluconeogenesis as diagrammed below occurs in two parts: a network of reactions converts mitochondrial pyruvate to cytosolic phosphoenolpyruvate; then phosphoenolpyruvate is converted to glucose 6-phosphate in a single sequence of cytosolic reactions. Three variants of the first part of the process are physiologically important. 1) A series of transport and transamination reactions convert mitochondrial oxaloacetate to cytosolic oxaloacetate which is converted to phosphoenolpyruvate by a hormonally regulated, cytosolic isoform of phosphoenolpyruvate carboxykinase. This variant allows regulated glucose synthesis from lactate. 2) Mitochondrial oxaloacetate is reduced to malate, which is exported to the cytosol and re-oxidized to oxaloacetate. This variant provides reducing equivalents to the cytosol, needed for glucose synthesis from amino acids such as alanine and glutamine. 3) Constitutively expressed mitochondrial phosphoenolpyruvate carboxykinase catalyzes the conversion of mitochondrial oxaloacetate to phosphoenolpyruvate which is then transported to the cytosol. The exact path followed by any one molecule of pyruvate through this reaction network is determined by the tissue in which the reactions are occurring, the source of the pyruvate, and the physiological stress that triggered gluconeogenesis. In all cases, the synthesis of glucose from two molecules of pyruvate requires the generation and consumption of two reducing equivalents as cytosolic NADH + H+. For pyruvate derived from lactate (variants 1 and 3), NADH + H+ is generated with the oxidation of lactate to pyruvate in the cytosol (a reaction of pyruvate metabolism not shown in the diagram). For pyruvate derived from amino acids (variant 2), mitochondrial NADH + H+ generated by glutamate dehydrogenase (a reaction of amino acid metabolism, not shown) is used to reduce oxaloacetate to malate, which is transported to the cytosol and re-oxidized, generating cytosolic NADH + H+. The synthesis of glucose from pyruvate also requires the consumption of six high-energy phosphates, four from ATP and two from GTP. In the second part of gluconeogenesis, cytosolic phosphoenolpyruvate, however derived, is converted to fructose 1,6-bisphosphate by reactions that are the reverse of steps of glycolysis. Hydrolysis of fructose 1,6-bisphosphate to fructose 6-phosphate is catalyzed by fructose 1,6-bisphosphatase, and fructose 6-phosphate is reversibly isomerized to glucose 6-phosphate |
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| AKR1B1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| ALDOC | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| ARL4D | Two-hybrid, two hybrid pooling approach | physical, physical association | 16169070, (Europe PMC) | 0.37 | BioGRID, IntAct |
| ASB16 | Affinity Capture-MS | physical | 24337577, (Europe PMC) | NA | BioGRID |
| ASB9 | Affinity Capture-MS | physical | 24337577, (Europe PMC) | NA | BioGRID |
| BRAF | Affinity Capture-MS | physical | 27034005, (Europe PMC) | NA | BioGRID |
| CEBPA | Affinity Capture-MS | physical | 18239623, (Europe PMC) | NA | BioGRID |
| CFTR | Affinity Capture-MS | physical | 26618866, (Europe PMC) | NA | BioGRID |
| CKMT1A | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| CKMT1B | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| CLNS1A | Co-fractionation | physical | 22939629, (Europe PMC) | NA | BioGRID |
| DLD | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DLST | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DNM1L | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DUSP13 | Affinity Capture-MS | physical | 27432908, (Europe PMC) | NA | BioGRID |
| ENO1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| ENO2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| ENO3 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| FABP5 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| G6PD | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| GAPDH | Co-fractionation | physical | 22939629, 26344197, (Europe PMC) | NA | BioGRID |
| GLO1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| HYPK | Affinity Capture-MS | physical | 23272104, (Europe PMC) | NA | BioGRID |
| IQCB1 | Affinity Capture-MS, anti tag coimmunoprecipitation | association, physical | 21565611, (Europe PMC) | 0.35 | BioGRID, IntAct |
| MDM2 | Affinity Capture-Western | physical | 24567357, (Europe PMC) | NA | BioGRID |
| NTRK1 | Affinity Capture-MS | physical | 25921289, (Europe PMC) | NA | BioGRID |
| NUTF2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PCGF1 | Affinity Capture-MS | physical | 26687479, (Europe PMC) | NA | BioGRID |
| PCNA | Affinity Capture-MS, Reconstituted Complex, far western blotting | direct interaction, physical, physical association | 20849852, (Europe PMC) | 0.54 | BioGRID, IntAct, MINT |
| PGAM2 | Affinity Capture-MS | physical | 26186194, 28514442, (Europe PMC) | NA | BioGRID |
| PHGDH | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PRCP | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PRDX2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PSMA3 | Affinity Capture-MS | physical | 22079093, (Europe PMC) | NA | BioGRID |
| RAB11A | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| RAB11B | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| SDHA | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| SIRT7 | Affinity Capture-MS | physical | 22586326, (Europe PMC) | NA | BioGRID |
| SOD1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TAGLN2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TPI1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TRIM25 | Affinity Capture-MS | physical | 29117863, (Europe PMC) | NA | BioGRID |
| TXN | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TYMS | Co-fractionation | physical | 22939629, (Europe PMC) | NA | BioGRID |
| UMPS | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| XRCC6 | Two-hybrid, two hybrid pooling approach | physical, physical association | 16169070, (Europe PMC) | 0.37 | BioGRID, IntAct |
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| ARL4D | Two-hybrid, two hybrid pooling approach | physical, physical association | 16169070, (Europe PMC) | 0.37 | BioGRID, IntAct |
| ATF2 | anti tag coimmunoprecipitation | association | 22304920, (Europe PMC) | 0.35 | IntAct |
| DCTN1 | two hybrid pooling approach | physical association | 16169070, (Europe PMC) | 0.37 | IntAct |
| DLD | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DLST | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DNM1L | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| IQCB1 | Affinity Capture-MS, anti tag coimmunoprecipitation | association, physical | 21565611, (Europe PMC) | 0.35 | BioGRID, IntAct |
| JUN | anti bait coimmunoprecipitation | association | 25609649, (Europe PMC) | 0.35 | IntAct, MINT |
| MAPK13 | anti tag coimmunoprecipitation | association | 19135240, (Europe PMC) | 0.35 | IntAct |
| PCNA | Affinity Capture-MS, Reconstituted Complex, far western blotting | direct interaction, physical, physical association | 20849852, (Europe PMC) | 0.54 | BioGRID, IntAct, MINT |
| PHOSPHO1 | pull down | association | 28330616, (Europe PMC) | 0.35 | IntAct |
| PKP4 | two hybrid pooling approach | physical association | 16169070, (Europe PMC) | 0.37 | IntAct |
| PPP1CA | anti bait coimmunoprecipitation | association | 25593058, (Europe PMC) | 0.35 | IntAct |
| SDHA | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| XRCC6 | Two-hybrid, two hybrid pooling approach | physical, physical association | 16169070, (Europe PMC) | 0.37 | BioGRID, IntAct |
| YWHAZ | pull down | physical association | 15161933, (Europe PMC) | 0.40 | IntAct, MINT |
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| PCNA | Affinity Capture-MS, Reconstituted Complex, far western blotting | direct interaction, physical, physical association | 20849852, (Europe PMC) | 0.54 | BioGRID, IntAct, MINT |
| YWHAZ | pull down | physical association | 15161933, (Europe PMC) | 0.40 | IntAct, MINT |
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| AKR1B1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| ALDOC | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| APP | anti bait coimmunoprecipitation | association | 16049941, (Europe PMC) | 0.35 | IntAct |
| ARL4D | Two-hybrid, two hybrid pooling approach | physical, physical association | 16169070, (Europe PMC) | 0.37 | BioGRID, IntAct |
| ASB16 | Affinity Capture-MS | physical | 24337577, (Europe PMC) | NA | BioGRID |
| ASB9 | Affinity Capture-MS | physical | 24337577, (Europe PMC) | NA | BioGRID |
| ATF2 | anti tag coimmunoprecipitation | association | 22304920, (Europe PMC) | 0.35 | IntAct |
| BRAF | Affinity Capture-MS | physical | 27034005, (Europe PMC) | NA | BioGRID |
| CEBPA | Affinity Capture-MS | physical | 18239623, (Europe PMC) | NA | BioGRID |
| CFTR | Affinity Capture-MS | physical | 26618866, (Europe PMC) | NA | BioGRID |
| CKMT1A | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| CKMT1B | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| CLNS1A | Co-fractionation | physical | 22939629, (Europe PMC) | NA | BioGRID |
| DCTN1 | two hybrid pooling approach | physical association | 16169070, (Europe PMC) | 0.37 | IntAct |
| DLD | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DLST | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DNM1L | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| DUSP13 | Affinity Capture-MS | physical | 27432908, (Europe PMC) | NA | BioGRID |
| ENO1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| ENO2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| ENO3 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| FABP5 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| G6PD | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| GAPDH | Co-fractionation | physical | 22939629, 26344197, (Europe PMC) | NA | BioGRID |
| GLO1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| HYPK | Affinity Capture-MS | physical | 23272104, (Europe PMC) | NA | BioGRID |
| IQCB1 | Affinity Capture-MS, anti tag coimmunoprecipitation | association, physical | 21565611, (Europe PMC) | 0.35 | BioGRID, IntAct |
| JUN | anti bait coimmunoprecipitation | association | 25609649, (Europe PMC) | 0.35 | IntAct, MINT |
| MAPK13 | anti tag coimmunoprecipitation | association | 19135240, (Europe PMC) | 0.35 | IntAct |
| MDM2 | Affinity Capture-Western | physical | 24567357, (Europe PMC) | NA | BioGRID |
| NTRK1 | Affinity Capture-MS | physical | 25921289, (Europe PMC) | NA | BioGRID |
| NUTF2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PCGF1 | Affinity Capture-MS | physical | 26687479, (Europe PMC) | NA | BioGRID |
| PCNA | Affinity Capture-MS, Reconstituted Complex, far western blotting | direct interaction, physical, physical association | 20849852, (Europe PMC) | 0.54 | BioGRID, IntAct, MINT |
| PGAM2 | Affinity Capture-MS | physical | 26186194, 28514442, (Europe PMC) | NA | BioGRID |
| PHGDH | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PHOSPHO1 | pull down | association | 28330616, (Europe PMC) | 0.35 | IntAct |
| PKP4 | two hybrid pooling approach | physical association | 16169070, (Europe PMC) | 0.37 | IntAct |
| PPP1CA | anti bait coimmunoprecipitation | association | 25593058, (Europe PMC) | 0.35 | IntAct |
| PRCP | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PRDX2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| PSMA3 | Affinity Capture-MS | physical | 22079093, (Europe PMC) | NA | BioGRID |
| RAB11A | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| RAB11B | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| SDHA | Affinity Capture-MS, cross-linking study | association, physical | 29128334, (Europe PMC) | 0.35 | BioGRID, IntAct |
| SIRT7 | Affinity Capture-MS | physical | 22586326, (Europe PMC) | NA | BioGRID |
| SOD1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TAGLN2 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TPI1 | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TRIM25 | Affinity Capture-MS | physical | 29117863, (Europe PMC) | NA | BioGRID |
| TXN | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| TYMS | Co-fractionation | physical | 22939629, (Europe PMC) | NA | BioGRID |
| UMPS | Co-fractionation | physical | 26344197, (Europe PMC) | NA | BioGRID |
| XRCC6 | Two-hybrid, two hybrid pooling approach | physical, physical association | 16169070, (Europe PMC) | 0.37 | BioGRID, IntAct |
| YWHAZ | pull down | physical association | 15161933, (Europe PMC) | 0.40 | IntAct, MINT |
| Phosphorylating Kinases | Phosphoresidues | Detection Method | PubMed IDs | Source |
| PAK1 | S118_QVKIWRRsYDVPPPP, S23_WNLENRFsGWYDADL, | LTP | 12189148,(Europe PMC) | PhosphoELM, PhosphoSitePlus, |
| Unknown | S118_QVKIWRRsYDVPPPP, S14_VLIRHGEsAWNLENR, S31_GWYDADLsPAGHEEA, T146_DRRYADLtEDQLPSC, Y119_VKIWRRSyDVPPPPM, Y133_MEPDHPFySNISKDR, Y26_ENRFSGWyDADLSPA, Y92_WRLNERHyGGLTGLN, | HTP, in vivo | 18083107, 18767875, 19651622, 20058876, 20068231, 20166139,(Europe PMC) | HPRD, PhosphoELM, |

