
- DEPOD Home
- About DEPOD
- User Manual
- Human Phosphatases
- Protein Substrates
- Non-Protein Substrates
- Pathway Mapping
- Dephosphorylation Network
- ID Mapping
- Download
- Acknowledgement
- Contact
- Imprint
- Links
- Ensembl
- Entrez Gene
- HGNC
- QuickGO
- Gene Expression Atlas
- OMIM
- UniProt
- IntEnz
- BRENDA
- LocDB
- M-CSA
- InterPro
- Pfam
- CATH
- Gene3D
- PDB
- PDBe
- DrugPort
- ModBase
- SwissModel
- ChEMBL
- Phospho.ELM
- PhosphoSitePlus
- NetworKIN
- HPRD
- PSICQUIC
- STRING
- IntAct
- MINT
- KEGG Pathway
- Reactome Pathway
- Pathway Commons
- PubMed
- ArchSchema
- Cytoscape Web
- Europe PMC
- EKPD
- SwissLipids
 Statistics
Statistics
241 human active and 13 inactive phosphatases in total;
194 phosphatases have substrate data;
--------------------------------
336 protein substrates;
83 non-protein substrates;
1215 dephosphorylation interactions;
--------------------------------
299 KEGG pathways;
876 Reactome pathways;
--------------------------------
last scientific update:
11 Mar, 2019
194 phosphatases have substrate data;
--------------------------------
336 protein substrates;
83 non-protein substrates;
1215 dephosphorylation interactions;
--------------------------------
299 KEGG pathways;
876 Reactome pathways;
--------------------------------
last scientific update:
11 Mar, 2019

Top
INPP5E
| Gene Name | INPP5E (QuickGO) | (A quick tutorial to explore the interctive visulaization) |
| Synonyms | INPP5E | |
| Protein Name | INPP5E |
|
| Alternative Name(s) |
72 kDa inositol polyphosphate 5-phosphatase;3.1.3.36;Phosphatidylinositol 4,5-bisphosphate 5-phosphatase;Phosphatidylinositol polyphosphate 5-phosphatase type IV; | |
| EntrezGene ID | 56623 (Comparitive Toxicogenomics) | |
| UniProt AC (Human) | Q9NRR6 (protein sequence) | |
| Enzyme Class | EC 3.1.3.36 (BRENDA | |
| Molecular Weight | 70205 Dalton | |
| Protein Length | 644 amino acids (AA) | |
| Genome Browsers | NCBI | ENSG00000148384 (Ensembl) | UCSC | |
| Crosslinking annotations | Query our ID-mapping table | |
| Orthologues | Quest For Orthologues (QFO) | GeneTree | |
| Classification | Superfamily: inositol-5-phosphatases (IP) | Historic class: Inositol-1,4,5-trisphosphate 5-phosphatase | CATH ID: 3.60.10.10 | SCOP Fold: DNase I | |
| Phosphatase activity | active | Catalytic signature motif: unknown | |
| Phosphorylation Network | Visualize | |
| Domain organization, Expression, Diseases | (show / hide) |
| Protein Domain | InterPro | Pfam | SMART | 3D Structure | PDB | DrugPort | ModBase | SwissModel |
| Polypeptide organization (Pfam) | No Pfam domain found | ||
| Catalytic Site | Mechanism and Catalytic Site Atlas | ||
| Gene Expression | Gene Expression Atlas | Diseases | OMIM | DisGeNet | ClinVar |
| Protein-protein interactions | STRING | IntAct | MINT | BioGRID | ||
| Phosphosites | Phospho.ELM | PhosphoSitePlus | ||
| Localization, Function, Catalytic activity and Sequence | (show / hide) |
| Localization (UniProt annotation) | |||
| Cytoplasm, cytoskeleton, cilium axoneme Golgi apparatus, Golgi stackmembrane Cell membrane Cell projection, ruffle Cytoplasm Note=Peripheral membrane proteinassociated with Golgi stacks | |||
| Function (UniProt annotation) | |||
| Converts phosphatidylinositol 3,4,5-trisphosphate(PtdIns 3,4,5-P3) to PtdIns-P2, and phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol 4-phosphate Specific forlipid substrates, inactive towards water soluble inositolphosphates | |||
| Catalytic Activity (UniProt annotation) | |||
| 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + H(2)O = 1-phosphatidyl-1D-myo-inositol 4-phosphate+ phosphate | |||
| Protein Sequence | |||
| MPSKAENLRPSEPAPQPPEGRTLQGQLPGAPPAQRAGSPPDAPGSESPALACSTPATPSGEDPPARAAPIAPRPPARPRL ERALSLDDKGWRRRRFRGSQEDLEARNGTSPSRGSVQSEGPGAPAHSCSPPCLSTSLQEIPKSRGVLSSERGSPSSGGNP LSGVASSSPNLPHRDAAVAGSSPRLPSLLPPRPPPALSLDIASDSLRTANKVDSDLADYKLRAQPLLVRAHSSLGPGRPR SPLACDDCSLRSAKSSFSLLAPIRSKDVRSRSYLEGSLLASGALLGADELARYFPDRNVALFVATWNMQGQKELPPSLDE FLLPAEADYAQDLYVIGVQEGCSDRREWETRLQETLGPHYVLLSSAAHGVLYMSLFIRRDLIWFCSEVECSTVTTRIVSQ IKTKGALGISFTFFGTSFLFITSHFTSGDGKVAERLLDYTRTVQALVLPRNVPDTNPYRSSAADVTTRFDEVFWFGDFNF RLSGGRTVVDALLCQGLVVDVPALLQHDQLIREMRKGSIFKGFQEPDIHFLPSYKFDIGKDTYDSTSKQRTPSYTDRVLY RSRHKGDICPVSYSSCPGIKTSDHRPVYGLFRVKVRPGRDNIPLAAGKFDRELYLLGIKRRISKEIQRQQALQSQNSSTI CSVS |
| Motif information from Eukaryotic Linear Motif atlas (ELM) | (show / hide) |
| Gene Ontology (P: Process; F: Function and C: Component terms) | (show / hide) | |
| GO:0001726 | C:ruffle |
| GO:0004439 | F:phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity |
| GO:0004445 | F:inositol-polyphosphate 5-phosphatase activity |
| GO:0005829 | C:cytosol |
| GO:0005886 | C:plasma membrane |
| GO:0005929 | C:cilium |
| GO:0005930 | C:axoneme |
| GO:0006661 | P:phosphatidylinositol biosynthetic process |
| GO:0032580 | C:Golgi cisterna membrane |
| GO:0046855 | P:inositol phosphate dephosphorylation |
| GO:0046856 | P:phosphatidylinositol dephosphorylation |
[Feedback|Contribution]
| Substrate ChEBI-ID | Substrate KEGG ID | DephosphoSite | Bioassay Type | PubMed ID | View in Europe PMC | Reliability of the interaction | Molecule |
| CHEBI:16595 | C01245 | D5 position | N/A | UniProt, | Europe PMC |  | 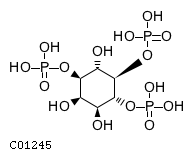 |
| CHEBI:16618 | C05981 | D5 position | In vitro | 10764818, | Europe PMC |  | 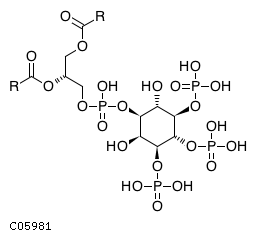 |
| CHEBI:16783 | C01272 | D5 position | N/A | UniProt, | Europe PMC |  | 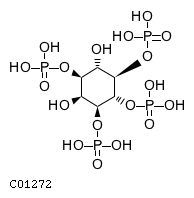 |
| CHEBI:18348 | C04637 | D5 position | In vitro | 10764818, | Europe PMC |  | 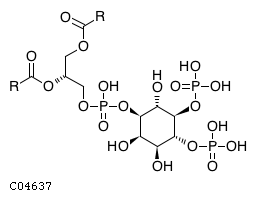 |
| Pathway ID | Pathway Name | Pathway Description (KEGG) |
| hsa00562 | Inositol phosphate metabolism | |
| hsa01100 | Metabolic pathways | |
| hsa04070 | Phosphatidylinositol signaling system | |
| Pathway ID | Pathway Name | Pathway Description (KEGG) |
| R-HSA-1660514 | Synthesis of PIPs at the Golgi membrane. | At the Golgi membrane, phosphatidylinositol 4-phosphate (PI4P) is primarily generated from phosphorylation of phosphatidylinositol (PI). Other phosphoinositides are also generated by the action of various kinases and phosphatases such as: phosphatidylinositol 3-phosphate (PI3P), phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2), phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) (Godi et al. 1999, Minogue et al. 2001, Rohde et al. 2003, Sbrissa et al. 2007, Sbrissa et al. 2008, Domin et al. 2000, Arcaro et al. 2000) |
| R-HSA-5624958 | ARL13B-mediated ciliary trafficking of INPP5E. | ARL13B is a ciliary-localized small GTPase with an atypical C-terminus containing a coiled coil domain and a proline rich domain (PRD) (Hori et al, 2008). Mutations in ARL13B are associated with the development of the ciliopathy Joubert's Syndrome (Cantagrel et al, 2008; Parisi et al, 2009). Studies in C. elegans and vertebrates suggest that ARL13B may play a role in stabilizing the interaction between the IFT A and B complexes and the kinesin-2 motors during anterograde traffic in the cilium (Cevik et al, 2010; Li et al, 2010; Cevik et al, 2013; reviewed in Li et al, 2012; Zhang et al, 2013). Recent work has shown an additional role for ARL13B in trafficking the inositol polyphosphate-5-phosphatase E (INPP5E) to the cilium through a network that also involves the phosphodiesterase PDE6D and the centriolar protein CEP164 (Humbert et al, 2012; Thomas et al, 2014; reviewed in Zhang et al, 2013). Mutations in INPP5E are also associated with the development of Joubert syndrome and other ciliopathies (Bielas et al, 2009; Jacoby et al, 2009; reviewed in Conduit et al, 2012) |
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| CBY1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CDC25C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CEP170 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CGN | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CWC25 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DCAF7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DCLK1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DENND1A | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| DENND4A | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DENND4C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| EIF4E2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| GIGYF1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| GIGYF2 | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| HDAC4 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| HDAC7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KCTD3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KIF13B | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KIF1C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KSR1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LIMA1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LPIN3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LRFN1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAGI1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAP3K21 | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| MAPKAP1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAST3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MTFR1L | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NADK | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NAV1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NF1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| OSBPL6 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PDE6D | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.51 | BioGRID, IntAct |
| PHLDB2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PLEKHA7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PPM1H | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PTPN13 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PTPN14 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RAB11FIP2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RALGPS2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RASAL2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RPGR | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.27, 0.35 | BioGRID, IntAct |
| RPTOR | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RTKN | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| SH3PXD2A | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SH3RF3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SIPA1L1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SRGAP2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| STARD13 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TANC2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TESK2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TIAM1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| YWHAE | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.51 | BioGRID, IntAct |
| ZBTB21 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| ZNF638 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| CDC25C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CEP170 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CGN | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CWC25 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DCAF7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DCLK1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DENND4A | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DENND4C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| EIF4E2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| GIGYF1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| HDAC4 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| HDAC7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KCTD3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KIF13B | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KIF1C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KSR1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LIMA1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LPIN3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LRFN1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAGI1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAPKAP1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAST3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MTFR1L | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NADK | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NAV1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NF1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| OSBPL6 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PDE6D | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.51 | BioGRID, IntAct |
| PHLDB2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PLEKHA7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PPM1H | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PTPN13 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PTPN14 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RAB11FIP2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RALGPS2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RASAL2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RPGR | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.27, 0.35 | BioGRID, IntAct |
| RPTOR | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SH3PXD2A | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SH3RF3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SIPA1L1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SRGAP2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| STARD13 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TANC2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TESK2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TIAM1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| YWHAE | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.51 | BioGRID, IntAct |
| ZBTB21 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| ZNF638 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
[Feedback|Contribution]
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| CBY1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CDC25C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CEP170 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CGN | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| CWC25 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DCAF7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DCLK1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DENND1A | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| DENND4A | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| DENND4C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| EIF4E2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| GIGYF1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| GIGYF2 | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| HDAC4 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| HDAC7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KCTD3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KIF13B | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KIF1C | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| KSR1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LIMA1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LPIN3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| LRFN1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAGI1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAP3K21 | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| MAPKAP1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MAST3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| MTFR1L | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NADK | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NAV1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| NF1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| OSBPL6 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PDE6D | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.51 | BioGRID, IntAct |
| PHLDB2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PLEKHA7 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PPM1H | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PTPN13 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| PTPN14 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RAB11FIP2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RALGPS2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RASAL2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RPGR | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.27, 0.35 | BioGRID, IntAct |
| RPTOR | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| RTKN | Affinity Capture-MS | physical | 27173435, (Europe PMC) | NA | BioGRID |
| SH3PXD2A | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SH3RF3 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SIPA1L1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| SRGAP2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| STARD13 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TANC2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TESK2 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| TIAM1 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| YWHAE | Affinity Capture-MS, inference by socio-affinity scoring, tandem affinity purification | association, physical, physical association | 27173435, , (Europe PMC) | 0.51 | BioGRID, IntAct |
| ZBTB21 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| ZNF638 | Affinity Capture-MS, inference by socio-affinity scoring | physical, physical association | 27173435, , (Europe PMC) | 0.27 | BioGRID, IntAct |
| Phosphorylating Kinases | Phosphoresidues | Detection Method | PubMed IDs | Source |
| Unknown | S99_RRRRFRGsQEDLEAR, T109_DLEARNGtSPSRGSV, | HTP, in vivo | 17287340, 17525332,(Europe PMC) | HPRD, PhosphoELM, |

