Top
IMPA1
Gene Name IMPA1 (QuickGO )Interactive visualization IMPA1 structures tutorial to explore the interctive visulaization)
Synonyms IMPA1, IMPA
Protein Name IMPA1
Alternative Name(s) Inositol monophosphatase 1;IMP 1;IMPase 1;3.1.3.25;D-galactose 1-phosphate phosphatase;3.1.3.94;Inositol-1(or 4)-monophosphatase 1;Lithium-sensitive myo-inositol monophosphatase A1;
EntrezGene ID 3612 (Comparitive Toxicogenomics)
UniProt AC (Human) P29218 (protein sequence )
Enzyme Class EC 3.1.3.25 3.1.3.94 (BRENDA Molecular Weight 30189 Dalton Protein Length 277 amino acids (AA) Genome Browsers NCBI | ENSG00000133731 (Ensembl) | UCSC Crosslinking annotations Query our ID-mapping tableOrthologues Quest For Orthologues (QFO) | GeneTree Classification Superfamily: adenosine_fructose_inositol-x-phosphatases (AFIxP) | Historic class: Inositol monophosphatase | CATH ID: 3.30.540.10, 3.40.190.80 | SCOP Fold: CP Phosphatase activity active | Catalytic signature motif: unknown Phosphorylation Network Visualize
Localization (UniProt annotation) Cytoplasm Function (UniProt annotation) Responsible for the provision of inositol required forsynthesis of phosphatidylinositol and polyphosphoinositides andhas been implicated as the pharmacological target for lithiumaction in brain Has broad substrate specificity and can use myo-inositol monophosphates, myo-inositol 1,3-diphosphate, myo-inositol 1,4-diphosphate, scyllo-inositol-phosphate, D-galactose1-phosphate, glucose-1-phosphate, glucose-6-phosphate, fructose-1-phosphate, beta-glycerophosphate, and 2'-AMP as substrates Catalytic Activity (UniProt annotation) Myo-inositol phosphate + H(2)O = myo-inositol+ phosphate Alpha-D-galactose 1-phosphate + H(2)O = D-galactose + phosphate Protein Sequence MADPWQECMDYAVTLARQAGEVVCEAIKNEMNVMLKSSPVDLVTATDQKVEKMLISSIKEKYPSHSFIGEESVAAGEKSI
LTDNPTWIIDPIDGTTNFVHRFPFVAVSIGFAVNKKIEFGVVYSCVEGKMYTARKGKGAFCNGQKLQVSQQEDITKSLLV
TELGSSRTPETVRMVLSNMEKLFCIPVHGIRSVGTAAVNMCLVATGGADAYYEMGIHCWDVAGAGIIVTEAGGVLMDVTG
GPFDLMSRRVIAANNRILAERIAKEIQVIPLQRDDED
ELM Motif Motif Instance Description Motif Regex NA NA NA NA
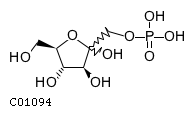
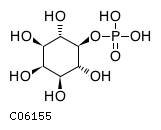
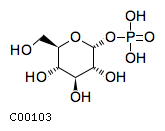
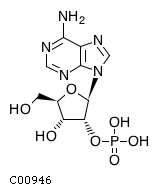
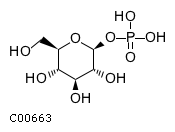
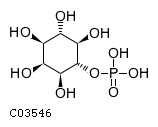
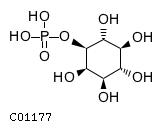
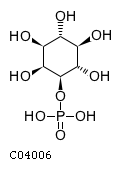
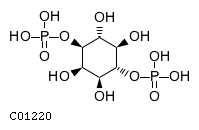
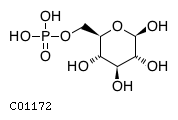
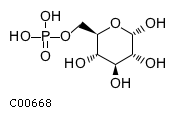
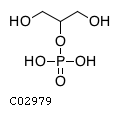
Substrate ChEBI-ID Substrate KEGG ID DephosphoSite Bioassay Type PubMed ID View in Europe PMC Reliability of the interaction Molecule CHEBI:25446 NA D6 position In vitro 17068342 , Europe PMC No image available CHEBI:62383 NA D2 position In vitro 17068342 , Europe PMC No image available CHEBI:37515 C01094 C-1 position In vitro 17068342 , Europe PMC CHEBI:37493 C06155 D5 position In vitro 17068342 , Europe PMC CHEBI:29042 C00103 C-1 position In vitro 17068342 , Europe PMC CHEBI:28223 C00946 2' position In vitro 17068342 , Europe PMC CHEBI:26613 NA N/A In vitro 17068342 , Europe PMC No image available CHEBI:26613 NA N/A In vitro 17068342 , Europe PMC No image available CHEBI:26613 NA N/A In vitro 17068342 , Europe PMC No image available CHEBI:16218 C00663 C-1 position In vitro 17068342 , Europe PMC CHEBI:18384 C03546 D4 position In vitro 17068342 , Europe PMC CHEBI:18297 C01177 D1 position In vitro 17068342 , Europe PMC CHEBI:18169 C04006 D3 position In vitro 17068342 , Europe PMC CHEBI:18105 C01094 C-1 position In vitro 17068342 , Europe PMC CHEBI:17816 C01220 N/A In vitro 17068342 , Europe PMC CHEBI:17719 C01172 C-6 position In vitro 17068342 , Europe PMC CHEBI:17665 C00668 C-6 position In vitro 17068342 , Europe PMC CHEBI:17270 C02979 2-position In vitro 17068342 , Europe PMC
Pathway ID Pathway Name Pathway Description (KEGG) R-HSA-1855183 Synthesis of IP2, IP, and Ins in the cytosol. Inositol phosphates IP2, IP and the six-carbon cyclic alcohol inositol (Ins) are produced by various phosphatases and the inositol-3-phosphate synthase 1 (ISYNA1) (Ju et al. 2004, Ohnishi et al. 2007, Irvine & Schell 2001, Bunney & Katan 2010)
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source CALB1 Protein-peptide physical 12176979 , (Europe PMC )NA BioGRID CYCS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID DUT Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID EIF6 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ELAVL1 Affinity Capture-RNA physical 19322201 , (Europe PMC )NA BioGRID IMPA1 Two-hybrid, two hybrid, two hybrid array, two hybrid pooling approach, two hybrid prey pooling approach physical, physical association 16189514 , 19447967 , 21516116 , 25416956 , Europe PMC )0.49, 0.75 BioGRID, IntAct, MINT IMPA2 Co-fractionation, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , 26344197 , Europe PMC )0.49, 0.56 BioGRID, IntAct ISCU Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID NTRK1 Affinity Capture-MS physical 25921289 , (Europe PMC )NA BioGRID NUDT9 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID NXF1 Reconstituted Complex physical 21965294 , (Europe PMC )NA BioGRID PGLS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PHPT1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID S100B Biochemical Activity, Co-localization, Reconstituted Complex physical 19593677 , (Europe PMC )NA BioGRID SCPEP1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID TXN Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID UGDH Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source IMPA1 Two-hybrid, two hybrid, two hybrid array, two hybrid pooling approach, two hybrid prey pooling approach physical, physical association 16189514 , 19447967 , 21516116 , 25416956 , Europe PMC )0.49, 0.75 BioGRID, IntAct, MINT IMPA2 Co-fractionation, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , 26344197 , Europe PMC )0.49, 0.56 BioGRID, IntAct
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source IMPA1 Two-hybrid, two hybrid, two hybrid array, two hybrid pooling approach, two hybrid prey pooling approach physical, physical association 16189514 , 19447967 , 21516116 , 25416956 , Europe PMC )0.49, 0.75 BioGRID, IntAct, MINT
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source CALB1 Protein-peptide physical 12176979 , (Europe PMC )NA BioGRID CYCS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID DUT Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID E2F3 pull down association 22157815 , (Europe PMC )0.35 IntAct, MINT EIF6 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ELAVL1 Affinity Capture-RNA physical 19322201 , (Europe PMC )NA BioGRID IMPA1 Two-hybrid, two hybrid, two hybrid array, two hybrid pooling approach, two hybrid prey pooling approach physical, physical association 16189514 , 19447967 , 21516116 , 25416956 , Europe PMC )0.49, 0.75 BioGRID, IntAct, MINT IMPA2 Co-fractionation, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , 26344197 , Europe PMC )0.49, 0.56 BioGRID, IntAct ISCU Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID NTRK1 Affinity Capture-MS physical 25921289 , (Europe PMC )NA BioGRID NUDT9 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID NXF1 Reconstituted Complex physical 21965294 , (Europe PMC )NA BioGRID PGLS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PHPT1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID S100B Biochemical Activity, Co-localization, Reconstituted Complex physical 19593677 , (Europe PMC )NA BioGRID SCPEP1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID TXN Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID UGDH Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID