Top
PUDP
| Gene Name | PUDP (QuickGO) | (A quick tutorial to explore the interctive visulaization) |
| Synonyms | PUDP , DXF68S1E, FAM16AX, GS1, HDHD1, HDHD1A | |
| Protein Name | PUDP |
|
| Alternative Name(s) |
Pseudouridine-5'-phosphatase;3.1.3.96;Haloacid dehalogenase-like hydrolase domain-containing protein 1;Haloacid dehalogenase-like hydrolase domain-containing protein 1A;Protein GS1;Pseudouridine-5'-monophosphatase;5'-PsiMPase; | |
| EntrezGene ID | 8226 (Comparitive Toxicogenomics) | |
| UniProt AC (Human) | Q08623 (protein sequence) | |
| Enzyme Class | EC 3.1.3.96 (BRENDA | |
| Molecular Weight | 25249 Dalton | |
| Protein Length | 228 amino acids (AA) | |
| Genome Browsers | NCBI | ENSG00000130021 (Ensembl) | UCSC | |
| Crosslinking annotations | Query our ID-mapping table | |
| Orthologues | Quest For Orthologues (QFO) | GeneTree | |
| Classification | Superfamily: HAD | Historic class: HAD-like hydrolase | CATH ID: 3.40.50.1000 | SCOP Fold: HAD | |
| Phosphatase activity | active | Catalytic signature motif: unknown | |
| Phosphorylation Network | Visualize | |
| Domain organization, Expression, Diseases | (show / hide) |
| Protein Domain | InterPro | Pfam | SMART | 3D Structure | PDB | DrugPort | ModBase | SwissModel |
| Polypeptide organization (Pfam) | No Pfam domain found | ||
| Catalytic Site | Mechanism and Catalytic Site Atlas | ||
| Gene Expression | Gene Expression Atlas | Diseases | OMIM | ClinVar |
| Protein-protein interactions | STRING | IntAct | MINT | BioGRID | ||
| Phosphosites | Phospho.ELM | PhosphoSitePlus | ||
| Localization, Function, Catalytic activity and Sequence | (show / hide) |
| Localization (UniProt annotation) | |||
| N/A | |||
| Function (UniProt annotation) | |||
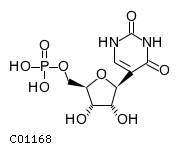
| Dephosphorylates pseudouridine 5'-phosphate, a potentialintermediate in rRNA degradation Pseudouridine is then excretedintact in urine | |||
| Catalytic Activity (UniProt annotation) | |||
| Pseudouridine 5'-phosphate + H(2)O =pseudouridine + phosphate | |||
| Protein Sequence | |||
| MAAPPQPVTHLIFDMDGLLLDTERLYSVVFQEICNRYDKKYSWDVKSLVMGKKALEAAQIIIDVLQLPMSKEELVEESQT KLKEVFPTAALMPGAEKLIIHLRKHGIPFALATSSGSASFDMKTSRHKEFFSLFSHIVLGDDPEVQHGKPDPDIFLACAK RFSPPPAMEKCLVFEDAPNGVEAALAAGMQVVMVPDGNLSRDLTTKATLVLNSLQDFQPELFGLPSYE |
| Motif information from Eukaryotic Linear Motif atlas (ELM) | (show / hide) |
| Gene Ontology (P: Process; F: Function and C: Component terms) | (show / hide) | |
| GO:0005737 | C:cytoplasm |
| GO:0005829 | C:cytosol |
| GO:0009117 | P:nucleotide metabolic process |
| GO:0016787 | F:hydrolase activity |
| GO:0016791 | F:phosphatase activity |
| GO:0043097 | P:pyrimidine nucleoside salvage |
| GO:0046872 | F:metal ion binding |
| GO:1990738 | F:pseudouridine 5'-phosphatase activity |