
Top
KEGG Pathway - Taste transduction - Homo sapiens (human) 
 | Phosphatases regulating this pathway |
Phosphatases regulating this pathway are unknown (yet)
 | Protein substrates in this pathway |
 | Non-protein substrates in this pathway |
| ChEBI ID | KEGG ID | ChEBIasciiName | Molecule | Dephosphorylated by: |
| CHEBI:16027 | C00020 | adenosine 5'-monophosphate | 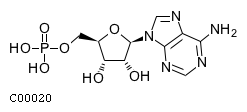 | ACPP, ALPL, NT5E, NT5C1A, |
| CHEBI:17202 | C00130 | IMP | 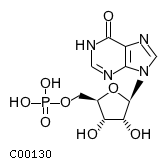 | NT5E, NT5C1A, NT5C2, NT5C, |
| CHEBI:17345 | C00144 | guanosine 5'-monophosphate | 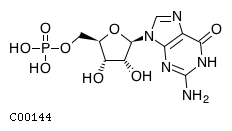 | NT5E, NT5C1A, NT5C2, NT5C, |
| CHEBI:16595 | C01245 | 1D-myo-inositol 1,4,5-trisphosphate | 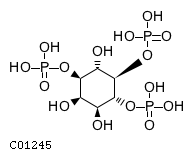 | INPP5B, INPP5A, INPP5E, INPP5J, INPP5K, OCRL, SYNJ1, SYNJ2, |

