Top
NT5C2
Localization (UniProt annotation) Cytoplasm Function (UniProt annotation) May have a critical role in the maintenance of aconstant composition of intracellular purine/pyrimidinenucleotides in cooperation with other nucleotidasesPreferentially hydrolyzes inosine 5'-monophosphate (IMP) and otherpurine nucleotides Catalytic Activity (UniProt annotation) A 5'-ribonucleotide + H(2)O = a ribonucleoside+ phosphate Protein Sequence MSTSWSDRLQNAADMPANMDKHALKKYRREAYHRVFVNRSLAMEKIKCFGFDMDYTLAVYKSPEYESLGFELTVERLVSI
GYPQELLSFAYDSTFPTRGLVFDTLYGNLLKVDAYGNLLVCAHGFNFIRGPETREQYPNKFIQRDDTERFYILNTLFNLP
ETYLLACLVDFFTNCPRYTSCETGFKDGDLFMSYRSMFQDVRDAVDWVHYKGSLKEKTVENLEKYVVKDGKLPLLLSRMK
EVGKVFLATNSDYKYTDKIMTYLFDFPHGPKPGSSHRPWQSYFDLILVDARKPLFFGEGTVLRQVDTKTGKLKIGTYTGP
LQHGIVYSGGSSDTICDLLGAKGKDILYIGDHIFGDILKSKKRQGWRTFLVIPELAQELHVWTDKSSLFEELQSLDIFLA
ELYKHLDSSSNERPDISSIQRRIKKVTHDMDMCYGMMGSLFRSGSRQTLFASQVMRYADLYAASFINLLYYPFSYLFRAA
HVLMPHESTVEHTHVDINEMESPLATRNRTSVDFKDTDYKRHQLTRSISEIKPPNLFPLAPQEITHCHDEDDDEEEEEEE
E
ELM Motif Motif Instance Description Motif Regex NA NA NA NA
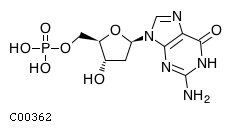
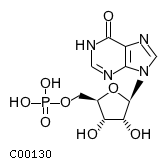
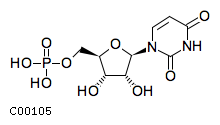
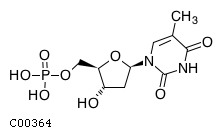
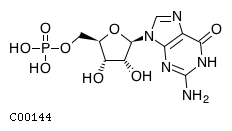
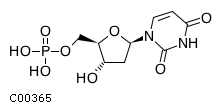
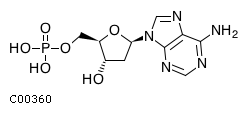
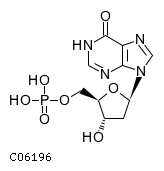
Substrate ChEBI-ID Substrate KEGG ID DephosphoSite Bioassay Type PubMed ID View in Europe PMC Reliability of the interaction Molecule CHEBI:16192 C00362 5' position In vitro 12907246 , 2848805 , Europe PMC CHEBI:17202 C00130 5' position In vitro 12907246 , 2848805 , Europe PMC CHEBI:16695 C00105 5' position In vitro 2848805 , Europe PMC CHEBI:17013 C00364 5' position In vitro 12907246 , Europe PMC CHEBI:17345 C00144 5' position In vitro 2848805 , Europe PMC CHEBI:17622 C00365 5' position In vitro 12907246 , Europe PMC CHEBI:17713 C00360 5' position In vitro 12907246 , Europe PMC CHEBI:28806 C06196 5' position In vitro 2848805 , Europe PMC
Pathway ID Pathway Name Pathway Description (KEGG) R-HSA-2161541 Abacavir metabolism. Abacavir activation proceeds steps of phosphorylation, deamination to yield carbovir monophosphate, and phosphorylation of the latter compound to yield the triphosphate. In addition, abacavir can be conjugated with glucuronide or oxidized to its 5'-carboxylate derivative, the two major forms in which it is excreted from the body (Yuen et al. 2008) R-HSA-74259 Purine catabolism. The purine bases guanine and hypoxanthine (derived from adenine by events in the purine salvage pathways) are converted to xanthine and then to uric acid, which is excreted from the body (Watts 1974). The end-point of this pathway in humans and hominoid primates is unusual. Most other mammals metabolize uric acid further to yield more soluble end products, and much speculation has centered on possible roles for high uric acid levels in normal human physiology
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source API5 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID ASB2 Affinity Capture-MS physical 24337577 , (Europe PMC )NA BioGRID ATP5PO Two-hybrid physical 25416956 , (Europe PMC )NA BioGRID BTF3L4 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID CALM1 Reconstituted Complex physical 15840729 , (Europe PMC )NA BioGRID CEP19 Proximity Label-MS, proximity-dependent biotin identification association, physical 26638075 , (Europe PMC )0.35 BioGRID, IntAct DRG2 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID DUSP3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID EHD4 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID FBF1 Proximity Label-MS, proximity-dependent biotin identification association, physical 26638075 , (Europe PMC )0.35 BioGRID, IntAct FXR2 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 19060904 , (Europe PMC )0.62 BioGRID, IntAct GNPDA1 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID HERC4 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID HNRNPL Affinity Capture-RNA physical 28611215 , (Europe PMC )NA BioGRID HSPBP1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID MOB1B Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct MOB3B Two-hybrid physical 16189514 , 19060904 , (Europe PMC )NA BioGRID MOB3C Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct NME7 Two-hybrid, two hybrid array, two hybrid pooling approach, two hybrid prey pooling approach, validated two hybrid physical, physical association 16189514 , 25416956 , (Europe PMC )0.67 BioGRID, IntAct NOC4L Two-hybrid, two hybrid pooling approach physical, physical association 16189514 , (Europe PMC )0.37 BioGRID, IntAct NT5C2 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 25416956 , (Europe PMC )0.55 BioGRID, IntAct NTRK1 Affinity Capture-MS physical 25921289 , (Europe PMC )NA BioGRID NUDT18 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 19060904 , (Europe PMC )0.62 BioGRID, IntAct PTGES3 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID RAD23A Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID SDCBP Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct SNX15 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID SZRD1 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID TRIM25 Affinity Capture-RNA physical 29117863 , (Europe PMC )NA BioGRID USP5 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID WARS Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source CEP19 Proximity Label-MS, proximity-dependent biotin identification association, physical 26638075 , (Europe PMC )0.35 BioGRID, IntAct FBF1 Proximity Label-MS, proximity-dependent biotin identification association, physical 26638075 , (Europe PMC )0.35 BioGRID, IntAct FXR2 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 19060904 , (Europe PMC )0.62 BioGRID, IntAct MOB1B Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct MOB3B two hybrid array, two hybrid pooling approach physical association 16189514 , 19060904 , (Europe PMC )0.62 IntAct MOB3C Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct NME7 Two-hybrid, two hybrid array, two hybrid pooling approach, two hybrid prey pooling approach, validated two hybrid physical, physical association 16189514 , 25416956 , (Europe PMC )0.67 BioGRID, IntAct NOC4L Two-hybrid, two hybrid pooling approach physical, physical association 16189514 , (Europe PMC )0.37 BioGRID, IntAct NT5C2 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 25416956 , (Europe PMC )0.55 BioGRID, IntAct NUDT18 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 19060904 , (Europe PMC )0.62 BioGRID, IntAct SDCBP Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct TP53 anti tag coimmunoprecipitation association 22653443 , (Europe PMC )0.35 IntAct, MINT
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source API5 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID ASB2 Affinity Capture-MS physical 24337577 , (Europe PMC )NA BioGRID ATP5O two hybrid array, two hybrid prey pooling approach, validated two hybrid physical association 25416956 , (Europe PMC )0.56 IntAct ATP5PO Two-hybrid physical 25416956 , (Europe PMC )NA BioGRID BTF3L4 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID CALM1 Reconstituted Complex physical 15840729 , (Europe PMC )NA BioGRID CEP19 Proximity Label-MS, proximity-dependent biotin identification association, physical 26638075 , (Europe PMC )0.35 BioGRID, IntAct DRG2 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID DUSP3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID EHD4 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID FBF1 Proximity Label-MS, proximity-dependent biotin identification association, physical 26638075 , (Europe PMC )0.35 BioGRID, IntAct FXR2 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 19060904 , (Europe PMC )0.62 BioGRID, IntAct GNPDA1 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID HERC4 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID HNRNPL Affinity Capture-RNA physical 28611215 , (Europe PMC )NA BioGRID HSPBP1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID MOB1B Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct MOB3B Two-hybrid physical 16189514 , 19060904 , (Europe PMC )NA BioGRID MOB3B two hybrid array, two hybrid pooling approach physical association 16189514 , 19060904 , (Europe PMC )0.62 IntAct MOB3C Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct NME7 Two-hybrid, two hybrid array, two hybrid pooling approach, two hybrid prey pooling approach, validated two hybrid physical, physical association 16189514 , 25416956 , (Europe PMC )0.67 BioGRID, IntAct NOC4L Two-hybrid, two hybrid pooling approach physical, physical association 16189514 , (Europe PMC )0.37 BioGRID, IntAct NT5C2 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 25416956 , (Europe PMC )0.55 BioGRID, IntAct NTRK1 Affinity Capture-MS physical 25921289 , (Europe PMC )NA BioGRID NUDT18 Two-hybrid, two hybrid array, two hybrid pooling approach physical, physical association 16189514 , 19060904 , (Europe PMC )0.62 BioGRID, IntAct PTGES3 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID RAD23A Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID SDCBP Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , (Europe PMC )0.56 BioGRID, IntAct SNX15 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID SZRD1 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID TP53 anti tag coimmunoprecipitation association 22653443 , (Europe PMC )0.35 IntAct, MINT TRIM25 Affinity Capture-RNA physical 29117863 , (Europe PMC )NA BioGRID USP5 Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID WARS Co-fractionation physical 22863883 , (Europe PMC )NA BioGRID
Phosphorylating Kinases Phosphoresidues Detection Method PubMed IDs Source
Visualize a network of protein kinases and phosphatases phosphorylating and dephosphorylating this protein respectively
Unknown S502_VDINEMEsPLATRNR , S527_KRHQLTRsISEIKPP , S529_HQLTRSIsEIKPPNL , Y60_MDYTLAVyKSPEYES , HTP, in vivo 18669648 , 20058876 , 20068231 ,(Europe PMC )HPRD, PhosphoELM ,