
CHEBI:16695
| Name | uridine 5'-monophosphate | 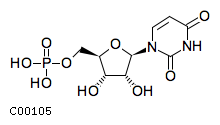 Download: mol | sdf |
| Synonyms | 05'-o-phosphonatouridine; 5'-ump; 5'-uridylate; 5'uridylic acid; Pu; Ump; Uridine 5'-(dihydrogen phosphate); Uridine 5'-monophosphate; Uridine 5'-phosphate; Uridine 5'-phosphoric acid; Uridine monophosphate; Uridine-5'-monophosphate; Uridylate; Uridylic acid; | |
| Definition | A pyrimidine ribonucleoside 5'-monophosphate having uracil as the nucleobase. | |
| Molecular Weight (Exact mass) | 324.1813 (324.0359) | |
| Molecular Formula | C9H13N2O9P | |
| SMILES | O[C@@H]1[C@@H](COP(O)(O)=O)O[C@H]([C@@H]1O)n1ccc(=O)[nH]c1=O | |
| InChI | InChI=1S/C9H13N2O9P/c12-5-1-2-11(9(15)10-5)8-7(14)6(13)4(20-8)3-19-21(16,17)18/h1-2,4,6-8,13-14H,3H2,(H,10,12,15)(H2,16,17,18)/t4-,6-,7-,8-/m1/s1 | |
| InChI Key | DJJCXFVJDGTHFX-XVFCMESISA-N | |
| Crosslinking annotations | KEGG:C00105 | 3DMET:B01161 | CAS:58-97-9 | ChEBI:16695 | ChEMBL:CHEMBL214393 | KNApSAcK:C00007311 | NIKKAJI:J4.594B | PDB-CCD:U | PDB-CCD:U5P | PubChem:3405 | |
| Pathway ID | Pathway Name | Pathway Description (KEGG) |
| map00240 | Pyrimidine metabolism | NA |
| map01100 | Metabolic pathways | NA |

