Top IMPA2
Localization (UniProt annotation) N/A Function (UniProt annotation) Can use myo-inositol monophosphates, scylloinositol 1,4-diphosphate, glucose-1-phosphate, beta-glycerophosphate, and 2'-AMP as substrates Has been implicated as the pharmacologicaltarget for lithium Li(+) action in brain Catalytic Activity (UniProt annotation) Myo-inositol phosphate + H(2)O = myo-inositol+ phosphate Protein Sequence MKPSGEDQAALAAGPWEECFQAAVQLALRAGQIIRKALTEEKRVSTKTSAADLVTETDHLVEDLIISELRERFPSHRFIA
EEAAASGAKCVLTHSPTWIIDPIDGTCNFVHRFPTVAVSIGFAVRQELEFGVIYHCTEERLYTGRRGRGAFCNGQRLRVS
GETDLSKALVLTEIGPKRDPATLKLFLSNMERLLHAKAHGVRVIGSSTLALCHLASGAADAYYQFGLHCWDLAAATVIIR
EAGGIVIDTSGGPLDLMACRVVAASTREMAMLIAQALQTINYGRDDEK
ELM Motif Motif Instance Description Motif Regex NA NA NA NA
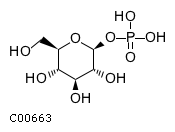
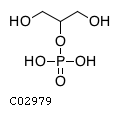
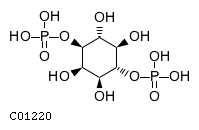
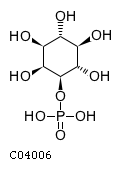
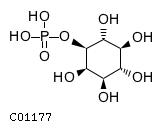
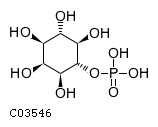
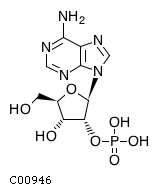
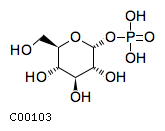
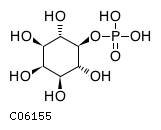
Substrate ChEBI-ID Substrate KEGG ID DephosphoSite Bioassay Type PubMed ID View in Europe PMC Reliability of the interaction Molecule CHEBI:16218 C00663 C-1 position In vitro 17068342 , Europe PMC CHEBI:17270 C02979 2-position In vitro 17068342 , Europe PMC CHEBI:17816 C01220 N/A In vitro 17068342 , Europe PMC CHEBI:18169 C04006 D3 position In vitro 17068342 , Europe PMC CHEBI:18297 C01177 D1 position In vitro 17068342 , Europe PMC CHEBI:18384 C03546 D4 position In vitro 17068342 , Europe PMC CHEBI:25446 NA D6 position In vitro 17068342 , Europe PMC No image available CHEBI:26613 NA N/A In vitro 17068342 , Europe PMC No image available CHEBI:28223 C00946 2' position In vitro 17068342 , Europe PMC CHEBI:29042 C00103 C-1 position In vitro 17068342 , Europe PMC CHEBI:37493 C06155 D5 position In vitro 17068342 , Europe PMC CHEBI:62383 NA D2 position In vitro 17068342 , Europe PMC No image available
Pathway ID Pathway Name Pathway Description (KEGG) R-HSA-1855183 Synthesis of IP2, IP, and Ins in the cytosol. Inositol phosphates IP2, IP and the six-carbon cyclic alcohol inositol (Ins) are produced by various phosphatases and the inositol-3-phosphate synthase 1 (ISYNA1) (Ju et al. 2004, Ohnishi et al. 2007, Irvine & Schell 2001, Bunney & Katan 2010)
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source AAR2 Affinity Capture-MS physical 28561026 , (Europe PMC )NA BioGRID ADSS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ALDOA Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ALDOC Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ARHGDIA Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID CARHSP1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID CYCS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID DUT Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID FSCN1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID GLO1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID GPX4 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID HNRNPL Affinity Capture-RNA physical 28611215 , (Europe PMC )NA BioGRID HSF2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID HSPE1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID IMPA1 Co-fractionation, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , 26344197 , Europe PMC )0.49, 0.56 BioGRID, IntAct IMPA2 Two-hybrid, two hybrid array physical, physical association 25416956 , (Europe PMC )0.37 BioGRID, IntAct ITPA Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID MSN Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID NPPA Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID NUDT9 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PABPC4 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PAICS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PARK7 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PDCD6 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PGLS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PLIN3 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID RBFOX1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID RDX Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID RPE Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID SCPEP1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID SPACA1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID STMN1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID TNFRSF14 Two-hybrid, two hybrid physical, physical association 21900206 , (Europe PMC )0.37 BioGRID, IntAct, MINT TRIM25 Affinity Capture-RNA physical 29117863 , (Europe PMC )NA BioGRID TXN Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID UBA3 Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , Europe PMC )0.70 BioGRID, IntAct ZIC1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source HERPUD1 two hybrid pooling approach physical association 16169070 , (Europe PMC )0.37 IntAct IMPA1 Co-fractionation, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , 26344197 , Europe PMC )0.49, 0.56 BioGRID, IntAct IMPA2 Two-hybrid, two hybrid array physical, physical association 25416956 , (Europe PMC )0.37 BioGRID, IntAct TNFRSF14 Two-hybrid, two hybrid physical, physical association 21900206 , (Europe PMC )0.37 BioGRID, IntAct, MINT UBA3 Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , Europe PMC )0.70 BioGRID, IntAct
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source AAR2 Affinity Capture-MS physical 28561026 , (Europe PMC )NA BioGRID ADSS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ALDOA Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ALDOC Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID ARHGDIA Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID CARHSP1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID CCT6A two hybrid fragment pooling approach physical association 23414517 , (Europe PMC )0.37 IntAct CYCS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID DUT Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID FSCN1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID GLO1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID GPX4 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID HERPUD1 two hybrid pooling approach physical association 16169070 , (Europe PMC )0.37 IntAct HNRNPL Affinity Capture-RNA physical 28611215 , (Europe PMC )NA BioGRID HSF2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID HSPE1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID IMPA1 Co-fractionation, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , 26344197 , Europe PMC )0.49, 0.56 BioGRID, IntAct IMPA2 Two-hybrid, two hybrid array physical, physical association 25416956 , (Europe PMC )0.37 BioGRID, IntAct ITPA Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID MSN Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID NPPA Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID NUDT9 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PABPC4 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PAICS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PARK7 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PDCD6 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PGLS Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID PLIN3 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID RBFOX1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID RDX Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID RPE Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID SCPEP1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID SPACA1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID STMN1 Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID TNFRSF14 Two-hybrid, two hybrid physical, physical association 21900206 , (Europe PMC )0.37 BioGRID, IntAct, MINT TRIM25 Affinity Capture-RNA physical 29117863 , (Europe PMC )NA BioGRID TXN Co-fractionation physical 26344197 , (Europe PMC )NA BioGRID UBA3 Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 25416956 , Europe PMC )0.70 BioGRID, IntAct ZIC1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID