
- DEPOD Home
- About DEPOD
- User Manual
- Human Phosphatases
- Protein Substrates
- Non-Protein Substrates
- Pathway Mapping
- Dephosphorylation Network
- ID Mapping
- Download
- Acknowledgement
- Contact
- Imprint
- Links
- Ensembl
- Entrez Gene
- HGNC
- QuickGO
- Gene Expression Atlas
- OMIM
- UniProt
- IntEnz
- BRENDA
- LocDB
- M-CSA
- InterPro
- Pfam
- CATH
- Gene3D
- PDB
- PDBe
- DrugPort
- ModBase
- SwissModel
- ChEMBL
- Phospho.ELM
- PhosphoSitePlus
- NetworKIN
- HPRD
- PSICQUIC
- STRING
- IntAct
- MINT
- KEGG Pathway
- Reactome Pathway
- Pathway Commons
- PubMed
- ArchSchema
- Cytoscape Web
- Europe PMC
- EKPD
- SwissLipids
 Statistics
Statistics
241 human active and 13 inactive phosphatases in total;
194 phosphatases have substrate data;
--------------------------------
336 protein substrates;
83 non-protein substrates;
1215 dephosphorylation interactions;
--------------------------------
299 KEGG pathways;
876 Reactome pathways;
--------------------------------
last scientific update:
11 Mar, 2019
194 phosphatases have substrate data;
--------------------------------
336 protein substrates;
83 non-protein substrates;
1215 dephosphorylation interactions;
--------------------------------
299 KEGG pathways;
876 Reactome pathways;
--------------------------------
last scientific update:
11 Mar, 2019

Top
PTP4A3
| Gene Name | PTP4A3 (QuickGO) | (A quick tutorial to explore the interctive visulaization) |
| Synonyms | PTP4A3, PRL3 | |
| Protein Name | PTP4A3 |
|
| Alternative Name(s) |
Protein tyrosine phosphatase type IVA 3;3.1.3.48;PRL-R;Protein-tyrosine phosphatase 4a3;Protein-tyrosine phosphatase of regenerating liver 3;PRL-3; | |
| EntrezGene ID | 11156 (Comparitive Toxicogenomics) | |
| UniProt AC (Human) | O75365 (protein sequence) | |
| Enzyme Class | EC 3.1.3.48 (BRENDA | |
| Molecular Weight | 19535 Dalton | |
| Protein Length | 173 amino acids (AA) | |
| Genome Browsers | NCBI | ENSG00000184489 (Ensembl) | ENSG00000275575 (Ensembl) | UCSC | |
| Crosslinking annotations | Query our ID-mapping table | |
| Orthologues | Quest For Orthologues (QFO) | GeneTree | |
| Classification | Superfamily: Class I Cys-based PTPs --> Family: DSP --> Subfamily: PRL | Historic class: Class I Cys-based PTPs --> VH1-like or DSPs --> PRLs | CATH ID: 3.90.190.10 | SCOP Fold: CC1 | |
| Phosphatase activity | active | Catalytic signature motif: HCVAGLGR | |
| Domain organization, Expression, Diseases | (show / hide) |
| Protein Domain | InterPro | Pfam | SMART | 3D Structure | PDB | DrugPort | ModBase | SwissModel |
| Polypeptide organization (Pfam) | No Pfam domain found | ||
| Catalytic Site | Mechanism and Catalytic Site Atlas | ||
| Gene Expression | Gene Expression Atlas | Diseases | OMIM | DisGeNet | ClinVar |
| Protein-protein interactions | STRING | IntAct | MINT | BioGRID | ||
| Phosphosites | Phospho.ELM | PhosphoSitePlus | ||
| Localization, Function, Catalytic activity and Sequence | (show / hide) |
| Localization (UniProt annotation) | |||
| Cell membrane Early endosome | |||
| Function (UniProt annotation) | |||
| Protein tyrosine phosphatase which stimulatesprogression from G1 into S phase during mitosis Enhances cellproliferation, cell motility and invasive activity, and promotescancer metastasis May be involved in the progression of cardiachypertrophy by inhibiting intracellular calcium mobilization inresponse to angiotensin II | |||
| Catalytic Activity (UniProt annotation) | |||
| Protein tyrosine phosphate + H(2)O = proteintyrosine + phosphate | |||
| Protein Sequence | |||
| MARMNRPAPVEVSYKHMRFLITHNPTNATLSTFIEDLKKYGATTVVRVCEVTYDKTPLEKDGITVVDWPFDDGAPPPGKV VEDWLSLVKAKFCEAPGSCVAVHCVAGLGRAPVLVALALIESGMKYEDAIQFIRQKRRGAINSKQLTYLEKYRPKQRLRF KDPHTHKTRCCVM |
| Motif information from Eukaryotic Linear Motif atlas (ELM) | (show / hide) |
| Gene Ontology (P: Process; F: Function and C: Component terms) | (show / hide) | |
| GO:0004727 | F:prenylated protein tyrosine phosphatase activity |
| GO:0005634 | C:nucleus |
| GO:0005737 | C:cytoplasm |
| GO:0005769 | C:early endosome |
| GO:0005886 | C:plasma membrane |
| GO:0006355 | P:regulation of transcription |
| GO:0007219 | DNA-templated |
| GO:0008138 | P:Notch signaling pathway |
| GO:0043117 | F:protein tyrosine/serine/threonine phosphatase activity |
| GO:0043542 | P:positive regulation of vascular permeability |
| GO:1900746 | P:endothelial cell migration |
| GO:1901224 | P:regulation of vascular endothelial growth factor signaling pathway |
| GO:1904951 | P:positive regulation of NIK/NF-kappaB signaling |
| GO:1990830 | P:positive regulation of establishment of protein localization |
Download FASTA sequences of PTP4A3-substrates in Humans
| Substrate Gene Name | Substrate UniProt_AC | DephosphoSite and sequence (+/-5) | Bioassay Type | PubMed ID | View in Europe PMC | Reliability of the interaction |
| EZR | P15311 | Thr-567_RDKYKtLRQIR | In vitro and in vivo | 18078820, | Europe PMC |   |
| SLC9A3R1 | O14745 | NA | In vivo | 25897829, | Europe PMC |  |
| ITGB1 | P05556 | NA | In vivo | 19930715, 16472776, | Europe PMC |  |
| KRT8 | P05787 | Ser-74_NQSLLsPLVLE, Ser-432 _YGGLTsPGLSY | In vivo | 19115206, | Europe PMC |  |
| STMN1 | P16949 | NA | In vivo | 20806969, | Europe PMC |  |
| NCL | P19338 | NA | In vivo | 20860603, | Europe PMC |  |
| BCAR1 | P56945 | NA | In vivo | 11355880, | Europe PMC |  |
| Substrate ChEBI-ID | Substrate KEGG ID | DephosphoSite | Bioassay Type | PubMed ID | View in Europe PMC | Reliability of the interaction | Molecule |
| CHEBI:18348 | C04637 | D5 position | In vitro | 21806020, | Europe PMC |  | 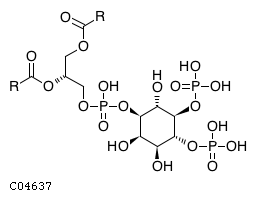 |
[ Feedback|Contribution]
[Feedback|Contribution]
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| AIFM1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATAD3A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ATP1B3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP2A2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CDK2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CDKN2A | Two-hybrid | physical | 27229929, (Europe PMC) | NA | BioGRID |
| CNNM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| DNAAF5 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ECPAS | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ELAVL1 | Affinity Capture-RNA | physical | 19322201, (Europe PMC) | NA | BioGRID |
| FANCI | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| GCN1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| GTF2I | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HEATR1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HNRNPM | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HUWE1 | Affinity Capture-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| IPO4 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| KIF1BP | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| LRPPRC | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MCM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MLF2 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| MMS19 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| MSH2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NCAPG | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NUP188 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PIK3R3 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| POLD1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PRPSAP1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSMD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PTP4A2 | Affinity Capture-MS, pull down | association, physical | 27432908, 28330616, 28514442, (Europe PMC) | 0.35 | BioGRID, IntAct |
| RAB3GAP2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SMAP1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| SMC2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SRPRB | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TMEM33 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TNPO2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TTC27 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| UBE4A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| USO1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| USP4 | Affinity Capture-Western | physical | 26669864, (Europe PMC) | NA | BioGRID |
| XPO5 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO7 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ABCD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| AIFM1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATAD3A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ATP1B3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP2A2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CDK2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CDKN2A | Two-hybrid | physical | 27229929, (Europe PMC) | NA | BioGRID |
| CNNM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| DNAAF5 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ECPAS | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ELAVL1 | Affinity Capture-RNA | physical | 19322201, (Europe PMC) | NA | BioGRID |
| FANCI | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| GCN1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| GTF2I | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HEATR1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HNRNPM | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HUWE1 | Affinity Capture-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| IPO4 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| KIF1BP | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| LRPPRC | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MCM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MLF2 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| MMS19 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| MSH2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NCAPG | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NUP188 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PIK3R3 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| POLD1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PRPSAP1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSMD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PTP4A2 | Affinity Capture-MS, pull down | association, physical | 27432908, 28330616, 28514442, (Europe PMC) | 0.35 | BioGRID, IntAct |
| RAB3GAP2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SMAP1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| SMC2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SRPRB | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TMEM33 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TNPO2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TTC27 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| UBE4A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| USO1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| USP4 | Affinity Capture-Western | physical | 26669864, (Europe PMC) | NA | BioGRID |
| XPO5 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO7 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| AHSA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| AIFM1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ARFGEF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATAD3A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1A4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1B3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP2A2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP6V1H | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATXN10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| BAG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CCAR2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CDK2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CHP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CLPX | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CNNM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CUL2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDOST | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDX20 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAAF5 {ECO:0000303|PubMed:25232951, | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJB11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJC11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DPM1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1054063 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058335 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058362 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058368 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058428 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058461 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058482 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058689 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ECPAS | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EFTUD2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EIF4G2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EPPK1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FANCI | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| FNTA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FNTB | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GBF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GCN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GEMIN4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GGT7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GTF2I | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HACD3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HEATR1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HEATR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HNRNPM | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HSD17B11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| IPO11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| IPO4 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| KIF1BP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| LRPPRC | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| LTN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MAD2L1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MCM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MCM7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MDN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MMS19 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MSH2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MYO1B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MYO1C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NCAPG | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NCDN | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUBP2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUP188 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NUP93 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OAT | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OPA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PCNA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDCD6IP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDS5A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDXDC1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| POLD1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PRPSAP1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSMD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSPH | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PTP4A2 | Affinity Capture-MS, pull down | association, physical | 27432908, 28330616, 28514442, (Europe PMC) | 0.35 | BioGRID, IntAct |
| PTP4A3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SEC61A1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SEC63 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SLC3A2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SMC2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SRPRB | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SSR1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STAT1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STRAP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SURF4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TANGO6 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TBC1D15 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TIMM23 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TMEM33 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TMEM41B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TNPO2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TNPO3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TOMM22 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TPM4 | proximity-dependent biotin identification, pull down | physical association | 28330616, (Europe PMC) | 0.52 | IntAct |
| TTC27 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TTI1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TUBG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE3C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE4A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UFL1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UNC45A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| USO1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO5 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO7 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ZW10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ABCD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| AHSA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| AIFM1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ARFGEF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATAD3A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1A4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1B3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP2A2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP6V1H | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATXN10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| BAG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CCAR2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CDK2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CHP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CLPX | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CNNM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CUL2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDOST | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDX20 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAAF5 {ECO:0000303|PubMed:25232951, | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJB11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJC11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DPM1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1054063 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058335 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058362 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058368 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058428 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058461 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058482 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058689 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ECPAS | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EFTUD2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EIF4G2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EPPK1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FANCI | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| FNTA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FNTB | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GBF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GCN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GEMIN4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GGT7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GTF2I | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HACD3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HEATR1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HEATR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HNRNPM | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HSD17B11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| IPO11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| IPO4 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| KIF1BP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| LRPPRC | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| LTN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MAD2L1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MCM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MCM7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MDN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MMS19 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MSH2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MYO1B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MYO1C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NCAPG | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NCDN | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUBP2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUP188 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NUP93 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OAT | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OPA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PCNA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDCD6IP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDS5A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDXDC1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| POLD1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PRPSAP1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSMD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSPH | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PTP4A2 | Affinity Capture-MS, pull down | association, physical | 27432908, 28330616, 28514442, (Europe PMC) | 0.35 | BioGRID, IntAct |
| PTP4A3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SEC61A1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SEC63 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SLC3A2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SMC2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SRPRB | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SSR1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STAT1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STRAP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SURF4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TANGO6 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TBC1D15 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TIMM23 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TMEM33 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TMEM41B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TNPO2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TNPO3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TOMM22 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TPM4 | proximity-dependent biotin identification, pull down | physical association | 28330616, (Europe PMC) | 0.52 | IntAct |
| TTC27 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TTI1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TUBG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE3C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE4A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UFL1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UNC45A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| USO1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO5 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO7 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ZW10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
[Feedback|Contribution]
| Interacting partner | Detection method | Interaction type | PubMed IDs | Interaction Score | Source |
| AHSA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| AIFM1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ARFGEF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATAD3A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ATAD3A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1A4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1B3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP2A2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP6V1H | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATXN10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| BAG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CCAR2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CDK2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CDKN2A | Two-hybrid | physical | 27229929, (Europe PMC) | NA | BioGRID |
| CHP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CLPX | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CNNM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CUL2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDOST | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDX20 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAAF5 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| DNAAF5 {ECO:0000303|PubMed:25232951, | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJB11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJC11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DPM1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1054063 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058335 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058362 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058368 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058428 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058461 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058482 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058689 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ECPAS | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ECPAS | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EFTUD2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EIF4G2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ELAVL1 | Affinity Capture-RNA | physical | 19322201, (Europe PMC) | NA | BioGRID |
| EPPK1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FANCI | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| FNTA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FNTB | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GBF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GCN1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| GCN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GEMIN4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GGT7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GTF2I | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HACD3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HEATR1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HEATR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HNRNPM | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HSD17B11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HUWE1 | Affinity Capture-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| IPO11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| IPO4 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| KIF1BP | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| KIF1BP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| LRPPRC | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| LTN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MAD2L1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MCM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MCM7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MDN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MLF2 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| MMS19 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| MMS19 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MSH2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MYO1B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MYO1C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NCAPG | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NCDN | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUBP2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUP188 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NUP93 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OAT | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OPA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PCNA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDCD6IP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDS5A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDXDC1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PIK3R3 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| POLD1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PRPSAP1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSMD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSPH | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PTP4A2 | Affinity Capture-MS, pull down | association, physical | 27432908, 28330616, 28514442, (Europe PMC) | 0.35 | BioGRID, IntAct |
| PTP4A3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SEC61A1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SEC63 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SLC3A2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SMAP1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| SMC2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SRPRB | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SSR1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STAT1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STRAP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SURF4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TANGO6 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TBC1D15 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TIMM23 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TMEM33 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TMEM41B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TNPO2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TNPO3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TOMM22 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TPM4 | proximity-dependent biotin identification, pull down | physical association | 28330616, (Europe PMC) | 0.52 | IntAct |
| TTC27 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TTI1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TUBG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE3C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE4A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| UBE4A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UFL1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UNC45A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| USO1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| USP4 | Affinity Capture-Western | physical | 26669864, (Europe PMC) | NA | BioGRID |
| XPO5 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO7 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ZW10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ABCD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| AHSA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| AIFM1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ARFGEF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATAD3A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ATAD3A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1A4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATP1B3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP2A2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ATP6V1H | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ATXN10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| BAG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CCAR2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CDK2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CDKN2A | Two-hybrid | physical | 27229929, (Europe PMC) | NA | BioGRID |
| CHP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CLPX | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| CNNM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| CUL2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDOST | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DDX20 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAAF5 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| DNAAF5 {ECO:0000303|PubMed:25232951, | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJB11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DNAJC11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| DPM1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1054063 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058335 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058362 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058368 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058428 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058461 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058482 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EBI-1058689 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ECPAS | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| ECPAS | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EFTUD2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| EIF4G2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| ELAVL1 | Affinity Capture-RNA | physical | 19322201, (Europe PMC) | NA | BioGRID |
| EPPK1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FANCI | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| FNTA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| FNTB | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GBF1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GCN1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| GCN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GEMIN4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GGT7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| GTF2I | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HACD3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HEATR1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HEATR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HNRNPM | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| HSD17B11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| HUWE1 | Affinity Capture-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| IPO11 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| IPO4 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| KIF1BP | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| KIF1BP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| LRPPRC | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| LTN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MAD2L1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MCM3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MCM7 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MDN1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MLF2 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| MMS19 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| MMS19 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MSH2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| MYO1B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| MYO1C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NCAPG | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NCDN | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUBP2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| NUP188 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| NUP93 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OAT | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| OPA1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PCNA | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDCD6IP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDS5A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PDXDC1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PIK3R3 | Proximity Label-MS | physical | 27880917, (Europe PMC) | NA | BioGRID |
| POLD1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PRPSAP1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSMD3 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| PSPH | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| PTP4A2 | Affinity Capture-MS, pull down | association, physical | 27432908, 28330616, 28514442, (Europe PMC) | 0.35 | BioGRID, IntAct |
| PTP4A3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| RAB3GAP2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SEC61A1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SEC63 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SLC3A2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SMAP1 | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| SMC2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SRPRB | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| SSR1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SSR4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STAT1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| STRAP | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| SURF4 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TANGO6 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TBC1D15 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TIMM23 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TMEM33 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TMEM41B | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TNPO2 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TNPO3 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TOMM22 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TPM4 | proximity-dependent biotin identification, pull down | physical association | 28330616, (Europe PMC) | 0.52 | IntAct |
| TTC27 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| TTI1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| TUBG2 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE3C | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UBE4A | Affinity Capture-MS | physical | 17353931, (Europe PMC) | NA | BioGRID |
| UBE4A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UFL1 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| UNC45A | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
| USO1 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| USP4 | Affinity Capture-Western | physical | 26669864, (Europe PMC) | NA | BioGRID |
| XPO5 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| XPO7 | Affinity Capture-MS, anti bait coimmunoprecipitation | physical, physical association | 17353931, (Europe PMC) | 0.40 | BioGRID, IntAct |
| ZW10 | anti bait coimmunoprecipitation | physical association | 17353931, (Europe PMC) | 0.40 | IntAct |
[Feedback|Contribution]

