Top
SGPP2
| Gene Name | SGPP2 (QuickGO) | (A quick tutorial to explore the interctive visulaization) |
| Synonyms | SGPP2 | |
| Protein Name | SGPP2 |
|
| Alternative Name(s) |
Sphingosine-1-phosphate phosphatase 2;SPPase2;Spp2;hSPP2;3.1.3.-;Sphingosine-1-phosphatase 2; | |
| EntrezGene ID | 130367 (Comparitive Toxicogenomics) | |
| UniProt AC (Human) | Q8IWX5 (protein sequence) | |
| Enzyme Class | N/A | |
| Molecular Weight | 44741 Dalton | |
| Protein Length | 399 amino acids (AA) | |
| Genome Browsers | NCBI | ENSG00000163082 (Ensembl) | UCSC | |
| Crosslinking annotations | Query our ID-mapping table | |
| Orthologues | Quest For Orthologues (QFO) | GeneTree | |
| Classification | Superfamily: glucose_lipid phosphatases | Historic class: Type 2 lipid phosphate phosphatase | CATH ID: 1.20.144.10 | SCOP Fold: Chloroperoxidase | |
| Phosphatase activity | active | Catalytic signature motif: unknown | |
| Domain organization, Expression, Diseases | (show / hide) |
| Protein Domain | InterPro | Pfam | SMART | 3D Structure | PDB | DrugPort | ModBase | SwissModel |
| Polypeptide organization (Pfam) | No Pfam domain found | ||
| Catalytic Site | Mechanism and Catalytic Site Atlas | ||
| Gene Expression | Gene Expression Atlas | Diseases | OMIM | DisGeNet | COSMIC | ClinVar |
| Protein-protein interactions | STRING | IntAct | MINT | BioGRID | ||
| Phosphosites | Phospho.ELM | PhosphoSitePlus | ||
| Localization, Function, Catalytic activity and Sequence | (show / hide) |
| Localization (UniProt annotation) | |||
| Endoplasmic reticulum membrane | |||
| Function (UniProt annotation) | |||
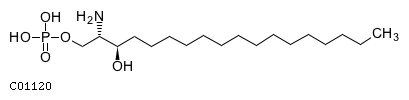
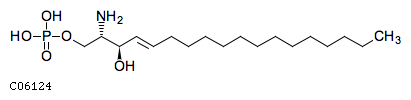
| Has specific phosphohydrolase activity towards sphingoidbase 1-phosphates Has high phosphohydrolase activity againstdihydrosphingosine-1-phosphate and sphingosine-1-phosphate (S1P)in vitro May play a role in attenuating intracellular sphingosine1-phosphate (S1P) signaling May play a role in pro-inflammatorysignaling | |||
| Catalytic Activity (UniProt annotation) | |||
| NA | |||
| Protein Sequence | |||
| MAELLRSLQDSQLVARFQRRCGLFPAPDEGPRENGADPTERAARVPGVEHLPAANGKGGEAPANGLRRAAAPEAYVQKYV VKNYFYYYLFQFSAALGQEVFYITFLPFTHWNIDPYLSRRLIIIWVLVMYIGQVAKDVLKWPRPSSPPVVKLEKRLIAEY GMPSTHAMAATAIAFTLLISTMDRYQYPFVLGLVMAVVFSTLVCLSRLYTGMHTVLDVLGGVLITALLIVLTYPAWTFID CLDSASPLFPVCVIVVPFFLCYNYPVSDYYSPTRADTTTILAAGAGVTIGFWINHFFQLVSKPAESLPVIQNIPPLTTYM LVLGLTKFAVGIVLILLVRQLVQNLSLQVLYSWFKVVTRNKEARRRLEIEVPYKFVTYTSVGICATTFVPMLHRFLGLP |
| Motif information from Eukaryotic Linear Motif atlas (ELM) | (show / hide) |
| Gene Ontology (P: Process; F: Function and C: Component terms) | (show / hide) | |
| GO:0005783 | C:endoplasmic reticulum |
| GO:0005789 | C:endoplasmic reticulum membrane |
| GO:0006670 | P:sphingosine metabolic process |
| GO:0016021 | C:integral component of membrane |
| GO:0030148 | P:sphingolipid biosynthetic process |
| GO:0042392 | F:sphingosine-1-phosphate phosphatase activity |
| GO:0070780 | F:dihydrosphingosine-1-phosphate phosphatase activity |