Top TPTE2
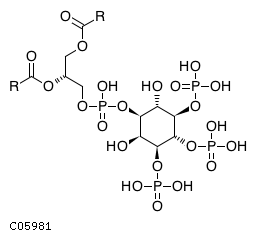
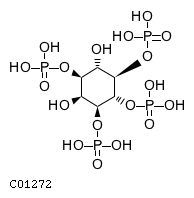
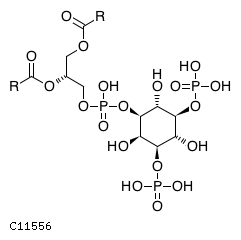
Pathway ID Pathway Name Pathway Description (KEGG) R-HSA-1660514 Synthesis of PIPs at the Golgi membrane. At the Golgi membrane, phosphatidylinositol 4-phosphate (PI4P) is primarily generated from phosphorylation of phosphatidylinositol (PI). Other phosphoinositides are also generated by the action of various kinases and phosphatases such as: phosphatidylinositol 3-phosphate (PI3P), phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2), phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) (Godi et al. 1999, Minogue et al. 2001, Rohde et al. 2003, Sbrissa et al. 2007, Sbrissa et al. 2008, Domin et al. 2000, Arcaro et al. 2000)
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source AKAP9 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID BRCA1 Two-hybrid physical 22990118 , (Europe PMC )NA BioGRID CCDC138 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC14 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC18 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC61 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC77 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CDK5RAP2 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CDK5RAP3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID CEP131 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CEP290 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CEP350 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CEP44 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID CEP72 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID DIAPH3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID DNAJB5 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID EPHA2 Two-hybrid physical 28065597 , (Europe PMC )NA BioGRID HAUS1 Affinity Capture-MS physical 27880917 , 28514442 , (Europe PMC )NA BioGRID HERC1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID IFFO1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID ITPRID2 Affinity Capture-MS, Proximity Label-MS physical 27880917 , 28514442 , (Europe PMC )NA BioGRID KIAA0232 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID KIAA0753 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID KIF1BP Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID KIF7 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID LGR4 Affinity Capture-MS physical 24639526 , (Europe PMC )NA BioGRID LMTK2 Two-hybrid physical 28065597 , (Europe PMC )NA BioGRID LYN Affinity Capture-Luminescence physical 23503679 , (Europe PMC )NA BioGRID MED4 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID MIB1 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID NCKAP5L Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID NUP133 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID OFD1 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID OTUD4 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID PCNT Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID PEX19 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID PIBF1 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID PKD2 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID PPP2R3A Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID PTK7 Two-hybrid physical 28065597 , (Europe PMC )NA BioGRID RHBDD1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID RMND1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID SH3GL1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID SSX2IP Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID STRN3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID STRN4 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TBC1D31 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID TEDC1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TFIP11 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TPTE Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID TRIM37 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID UBB Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source AKAP9 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID BRCA1 Two-hybrid physical 22990118 , (Europe PMC )NA BioGRID CCDC138 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC14 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC18 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC61 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CCDC77 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CDK5RAP2 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CDK5RAP3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID CEP131 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CEP290 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CEP350 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID CEP44 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID CEP72 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID DIAPH3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID DNAJB5 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID EPHA2 Two-hybrid physical 28065597 , (Europe PMC )NA BioGRID HAUS1 Affinity Capture-MS physical 27880917 , 28514442 , (Europe PMC )NA BioGRID HERC1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID IFFO1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID ITPRID2 Affinity Capture-MS, Proximity Label-MS physical 27880917 , 28514442 , (Europe PMC )NA BioGRID KIAA0232 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID KIAA0753 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID KIF1BP Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID KIF7 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID LGR4 Affinity Capture-MS physical 24639526 , (Europe PMC )NA BioGRID LMTK2 Two-hybrid physical 28065597 , (Europe PMC )NA BioGRID LYN Affinity Capture-Luminescence physical 23503679 , (Europe PMC )NA BioGRID MED4 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID MIB1 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID NCKAP5L Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID NUP133 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID OFD1 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID OTUD4 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID PCNT Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID PEX19 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID PIBF1 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID PKD2 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID PPP2R3A Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID PTK7 Two-hybrid physical 28065597 , (Europe PMC )NA BioGRID RHBDD1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID RMND1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID SH3GL1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID SSX2IP Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID STRN3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID STRN4 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TBC1D31 Proximity Label-MS physical 27880917 , (Europe PMC )NA BioGRID TEDC1 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TFIP11 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TPTE Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID TRIM37 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID UBB Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID