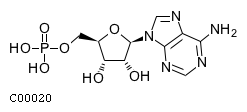
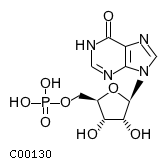
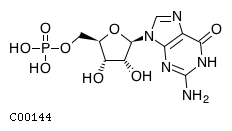
Top NT5E
Localization (UniProt annotation) Cell membrane; Lipid-anchor, GPI-anchor Function (UniProt annotation) Hydrolyzes extracellular nucleotides into membranepermeable nucleosides Exhibits AMP-, NAD-, and NMN-nucleosidaseactivities Catalytic Activity (UniProt annotation) A 5'-ribonucleotide + H(2)O = a ribonucleoside+ phosphate Protein Sequence MCPRAARAPATLLLALGAVLWPAAGAWELTILHTNDVHSRLEQTSEDSSKCVNASRCMGGVARLFTKVQQIRRAEPNVLL
LDAGDQYQGTIWFTVYKGAEVAHFMNALRYDAMALGNHEFDNGVEGLIEPLLKEAKFPILSANIKAKGPLASQISGLYLP
YKVLPVGDEVVGIVGYTSKETPFLSNPGTNLVFEDEITALQPEVDKLKTLNVNKIIALGHSGFEMDKLIAQKVRGVDVVV
GGHSNTFLYTGNPPSKEVPAGKYPFIVTSDDGRKVPVVQAYAFGKYLGYLKIEFDERGNVISSHGNPILLNSSIPEDPSI
KADINKWRIKLDNYSTQELGKTIVYLDGSSQSCRFRECNMGNLICDAMINNNLRHTDEMFWNHVSMCILNGGGIRSPIDE
RNNGTITWENLAAVLPFGGTFDLVQLKGSTLKKAFEHSVHRYGQSTGEFLQVGGIHVVYDLSRKPGDRVVKLDVLCTKCR
VPSYDPLKMDEVYKVILPNFLANGGDGFQMIKDELLRHDSGDQDINVVSTYISKMKVIYPAVEGRIKFSTGSHCHGSFSL
IFLSLWAVIFVLYQ
ELM Motif Motif Instance Description Motif Regex NA NA NA NA
Pathway ID Pathway Name Pathway Description (KEGG) R-HSA-196807 Nicotinate metabolism. Nicotinate (niacin) and nicotinamide are precursors of the coenzymes nicotinamide-adenine dinucleotide (NAD+) and nicotinamide-adenine dinucleotide phosphate (NADP+). When NAD+ and NADP+ are interchanged in a reaction with their reduced forms, NADH and NADPH respectively, they are important cofactors in several hundred redox reactions. Nicotinate is synthesized from 2-amino-3-carboxymuconate semialdehyde, an intermediate in the catabolism of the essential amino acid tryptophan (Magni et al. 2004) R-HSA-73621 Pyrimidine catabolism. In parallel sequences of three reactions each, thymine is converted to beta-aminoisobutyrate and uracil is converted to beta-alanine. Both of these molecules are excreted in human urine and appear to be normal end products of pyrimidine catabolism (Griffith 1986; Webster et al. 2001). Mitochondrial AGXT2, however, can also catalyze the transamination of both molecules with pyruvate, yielding 2-oxoacids that can be metabolized further by reactions of branched-chain amino acid and short-chain fatty acid catabolism (Tamaki et al. 2000). The importance of these reactions in normal human pyrimidine catabolism has not been well worked out R-HSA-74259 Purine catabolism. The purine bases guanine and hypoxanthine (derived from adenine by events in the purine salvage pathways) are converted to xanthine and then to uric acid, which is excreted from the body (Watts 1974). The end-point of this pathway in humans and hominoid primates is unusual. Most other mammals metabolize uric acid further to yield more soluble end products, and much speculation has centered on possible roles for high uric acid levels in normal human physiology
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source AFF1 Two-hybrid physical 19956800 , (Europe PMC )NA BioGRID ARF5 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID ARL2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID ARL8A Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID ATR Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID CHD1L Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID CXCR4 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID CYP2S1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID DEGS1 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID DNAAF5 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID FLVCR1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID GHITM Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID GJA1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID GLMN Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID GNL3L Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID HSDL1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID MTOR Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID NDUFA2 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID NR2F2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID PCDH7 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID PDS5A Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID PLAA Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID RAB9A Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID RDH13 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID RFC2 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID RHOG Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID RPTOR Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SAR1B Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SCAMP1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC16A2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC26A6 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC30A5 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID SLC41A1 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID SLC44A1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC47A1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC6A8 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC7A2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID STEAP3 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SYNJ2BP Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID TMEM177 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID TMEM192 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TNPO2 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID USP22 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID USP3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID USP36 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID VANGL2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID XPO7 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source AFF1 Two-hybrid physical 19956800 , (Europe PMC )NA BioGRID ARF5 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID ARL2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID ARL8A Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID ATR Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID CAT proximity-dependent biotin identification association 29568061 , (Europe PMC )0.35 IntAct CHD1L Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID CXCR4 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID CYP2S1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID DEGS1 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID DNAAF5 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID FLVCR1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID GHITM Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID GJA1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID GLMN Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID GNL3L Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID HSDL1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID MTOR Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID NDUFA2 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID NR2F2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID NT5E x-ray crystallography direct interaction 23142347 , (Europe PMC )0.44 IntAct PCDH7 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID PDS5A Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID PLAA Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID RAB11A proximity-dependent biotin identification association 29568061 , (Europe PMC )0.35 IntAct RAB9A Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID RDH13 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID RFC2 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID RHOG Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID RPTOR Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SAR1B Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SCAMP1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC16A2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC26A6 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC30A5 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID SLC41A1 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID SLC44A1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC47A1 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC6A8 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SLC7A2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID STEAP3 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID SYNJ2BP Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID TMEM177 Co-fractionation physical 22939629 , (Europe PMC )NA BioGRID TMEM192 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID TNPO2 Affinity Capture-MS physical 26186194 , (Europe PMC )NA BioGRID USP22 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID USP3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID USP36 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID VANGL2 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID XPO7 Affinity Capture-MS physical 26186194 , 28514442 , (Europe PMC )NA BioGRID