Top MTMR7
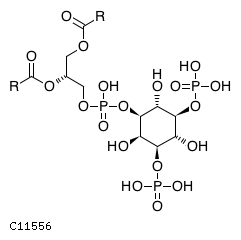
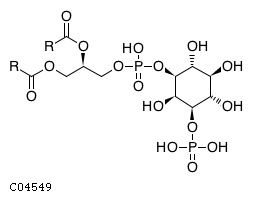
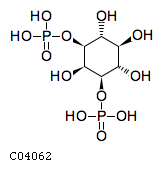
Pathway ID Pathway Name Pathway Description (KEGG) R-HSA-1660517 Synthesis of PIPs at the late endosome membrane. At the late endosome membrane, the primary event is the dephosphorylation of the phosphoinositide phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) to phosphatidylinositol 3-phosphate (PI3P) and phosphatidylinositol 5-phosphate (PI5P) (Sbrissa et al. 2007, Sbrissa et al. 2008, Cao et al. 2007, Cao et al. 2008, Arcaro et al. 2000, Kim et al. 2002) R-HSA-1855183 Synthesis of IP2, IP, and Ins in the cytosol. Inositol phosphates IP2, IP and the six-carbon cyclic alcohol inositol (Ins) are produced by various phosphatases and the inositol-3-phosphate synthase 1 (ISYNA1) (Ju et al. 2004, Ohnishi et al. 2007, Irvine & Schell 2001, Bunney & Katan 2010)
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source APOLD1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID AURKA Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CAMSAP3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CCDC6 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CCNH Affinity Capture-MS, two hybrid array, two hybrid prey pooling approach physical, physical association 27880917 , Europe PMC )0.49 BioGRID, IntAct CDC42BPB Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CORO1B Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CTBP1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CTBP2 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CTNND1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CYTH2 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CYTH3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID DOCK7 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID GNB2 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID GTF2I Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID HAUS1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID HNRNPC Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MIA3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MRPS34 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID MTHFD1L Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID MTMR6 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MTMR7 Affinity Capture-MS, Affinity Capture-Western physical 12890864 , 27432908 , (Europe PMC )NA BioGRID MTMR8 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MTMR9 Affinity Capture-MS, Affinity Capture-Western, Reconstituted Complex, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 12890864 , 25416956 , 27880917 , 28514442 , Europe PMC )0.70 BioGRID, IntAct PCM1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID RALGAPA2 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID RARS Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID RPS10P5 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID SAV1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID SOAT1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID STK3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID STXBP3 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID TSR3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID VIPAS39 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID VPS16 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID VPS33A Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID VPS33B Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID XRCC4 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID YEATS4 Affinity Capture-MS physical 27432908 , 27880917 , (Europe PMC )NA BioGRID
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source MTMR9 Affinity Capture-MS, Affinity Capture-Western, Reconstituted Complex, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 12890864 , 25416956 , 27880917 , 28514442 , Europe PMC )0.70 BioGRID, IntAct
Visualize Download
Interacting partner Detection method Interaction type PubMed IDs Interaction Score Source APOLD1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID AURKA Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CAMSAP3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CCDC6 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CCNH Affinity Capture-MS, two hybrid array, two hybrid prey pooling approach physical, physical association 27880917 , Europe PMC )0.49 BioGRID, IntAct CDC42BPB Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CORO1B Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CTBP1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CTBP2 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CTNND1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CYTH2 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID CYTH3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID DOCK7 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID GNB2 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID GTF2I Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID HAUS1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID HNRNPC Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MIA3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MRPS34 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID MTHFD1L Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID MTMR6 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MTMR7 Affinity Capture-MS, Affinity Capture-Western physical 12890864 , 27432908 , (Europe PMC )NA BioGRID MTMR8 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID MTMR9 Affinity Capture-MS, Affinity Capture-Western, Reconstituted Complex, Two-hybrid, two hybrid array, two hybrid prey pooling approach, validated two hybrid physical, physical association 12890864 , 25416956 , 27880917 , 28514442 , Europe PMC )0.70 BioGRID, IntAct PCM1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID RALGAPA2 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID RARS Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID RPS10P5 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID SAV1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID SOAT1 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID STK3 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID STXBP3 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID TSR3 Affinity Capture-MS physical 28514442 , (Europe PMC )NA BioGRID VIPAS39 Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID VPS16 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID VPS33A Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID VPS33B Affinity Capture-MS physical 27880917 , (Europe PMC )NA BioGRID XRCC4 Affinity Capture-MS physical 27432908 , (Europe PMC )NA BioGRID YEATS4 Affinity Capture-MS physical 27432908 , 27880917 , (Europe PMC )NA BioGRID